Abstract
16S rRNA gene sequencing was used to analyze the changes in the microbial community structure of Macrobrachium rosenbergii (M. rosenbergii) larvae and environmental water at different stages (0, 7, 14 and 21 days). The PICRUSt was used to precalculated the predicted genes and their functions. The Alpha and Beta diversity analysis showed that the abundance and uniformity of the flora of larvae and water samples were significantly different at four stages. At the phylum level, Proteobacteria, Planctomycetes, Firmicutes and Bacteroidetes were the dominant flora of larvae, while Proteobacteria, Bacteroidetes and Actinobacteria were the dominant flora of environmental water. But at the genus level, Raoultella and Blastopirellula were the main changes in larvae, and Dyadobacter, Flavobacterium and Pseudomonas were the main changes in environmental water. The gene function prediction analysis indicated that the amino acid metabolism, immune system, immune system disease, nervous system, sensory system and other pathways enriched in the different stages of larvae and environmental water were closely related to the breeding of M. rosenbergii. This research might provide theoretical basis and guidance for the high-quality breeding of M. rosenbergii.
HIGHLIGHTS
The abundance and uniformity of the flora of larvae and environmental water were significantly different at different stages.
Several flora were found that related to the growth and development of larval.
KEGG pathways enriched at different stages of larvae and environmental water were closely related to the breeding of larvae.
1. Introduction
Macrobrachium rosenbergii (M. rosenbergii) is a commercially important species of freshwater prawn, which is widely cultured all over the world. Because of its advantages, such as large size, delicious meat, rich nutrition, easy breeding and so on, it is well received by the market (Nguyen et al. 2019). China has been the largest industry of M. rosenbergii for more than 20 years worldwide. In China, the annual production of M. rosenbergii is nearly 150,000 tons, and the breeding volume is about 40 billion, which has made a huge contribution to the consumer market (Yang et al. Citation2012). Therefore, the scale of intensive farming of M. rosenbergii has continued to expand in recent years. However, due to high-density breeding and high-frequency feeding, the water quality of the nursery system deteriorates, which in turn induces larval disease, growth failure, and even death (Li et al. Citation2019). These problems have seriously affected the production and quality of shrimp larvae, and are not conducive to the healthy development of M. rosenbergii aquaculture (Wang and Liu Citation2017).
The characteristics of the microbial community in the nursery system and its directional regulation are the current research hotspots in the field of aquaculture. 16S rRNA gene sequencing is a high-throughput sequencing technology based on the microbial community structure in a specific environment as the research object (Madhavan et al. Citation2017). It analyzes the relationship on gene level between the diversity of the microbial population, the change of the population structure and the environment (Sudarikov et al. Citation2017; Potapov et al. Citation2019). Nowadays, this technology has been widely used to study the diversity of microorganisms in various environments such as soil, sewage, animals and humans (Laudadio et al. Citation2018; Liu et al. Citation2018; Lee et al. Citation2019; Shang et al. Citation2019; Zhang et al. Citation2019; Zhu et al. Citation2019). Through the study of the structure of the intestinal flora of M. rosenbergii, Dong et al. (Citation2019) found that the most dominant bacteria in different culture models were significantly different. Ni et al. (Citation2019) compared the microbial community structure of M. rosenbergii seedling rearing water with different salinities. Their results showed that the microbial community structure of rearing water were significantly different, and also found that low salinity could increase the possibility of a variety of potential pathogens. Other studies on M. rosenbergii flora mainly focus on the effects of feed probiotics on intestinal flora and healthy growth (Qin Citation2001; Li et al. Citation2002). However, the structural changes of M. rosenbergii larvae and water flora at different developmental stages have not yet been verified. Thus, it is very important to study the microbial community characteristics of M. rosenbergii larvae at different developmental stages for the health control of the later stage of seedling growth.
In this study, 16S rRNA gene high-throughput sequencing was used to analyze the changes of microbial community structure in M. rosenbergii larvae and environmental water at different developmental stages. The purpose of this research is to discover the dominant flora and functional pathways closely related to the breeding of larvae, and to provide a theoretical basis for the high-quality breeding of M. rosenbergii.
2. Materials and methods
2.1. Animal source and culture
The prawn population was established in the national M. rosenbergii seed multiplication farm, Huzhou, Zhejiang, China. All the prawns were reared in the same temperature ponds and fed the same diet. M. rosenbergii larvae were randomly sorted into the 9 similar size rectangular tanks (200 L capacity filled to 160 L). The artificial seawater for larvae has undergone chlorine treatment, disinfection, precipitation purification and other processes to keep the salinity at 5‰–6‰. The larvae were fed Brine Shrimp, a small amount of egg custard from the ninth day of age. The larvae were initially fed with Brine Shrimp, and were replaced by egg custard at the 7th day of age. The larvae were hand-fed two times daily (07:30, 18:00) at the same fixed rate. During the feeding trial, the water quality parameters were recorded as follows: temperature 29.0 ± 1.0 °C, dissolved oxygen 6.2 ± 0.7 mg/L, pH 7.7–8.2, density 100,000–200,000 per m3, light intensity 1000–1500 lux. All rearing tanks were provided with continuous aeration and maintained in natural photoperiod.
2.2. Sample collection
Each experimental group of samples collected included four various stages of development and environmental water samples, including 0, 7, 14 and 21 days (initial hatching state, bait replacement stage, larval metamorphosis stage, and environmental water salinity change stage, respectively) after larva hatching. In addition, three replicates were set for each group, and 10 prawns were randomly selected from each replicate group. The mixed whole larvae tissue sample was obtained from each replicate. The environmental water samples were collected as follow: after the air pump was turned off and the water body was still, then taken water samples using a beaker; after the larvae ware separated by a filter screen, collect the water samples into the water sample bag for standby. Fresh mixed tissue and environmental water samples were immediately placed in liquid nitrogen, and stored at −80 °C for DNA extraction after back to laboratory.
2.3. DNA isolation
Total DNA was extracted from the 12 tissue samples and 12 environmental water samples using an PowerSoli® DNA Isolation Kit (MOBIO, USA) according to the manufacturer’s instruction. The quantity and quality of DNA samples were measured with a NanoDrop2000/2000C Nucleic Acid Protein Detector and by 1% agarose gel electrophoresis. Then, the genomic DNA was diluted with ultra-pure water to a final concentration of 20 ng/ml and stored at 4 °C.
2.4. Sequencing and sequence analysis
The V3 and V4 hypervariable region of the 16S rRNA gene was amplified by PCR using the sample-specific sequence barcoded fusion primers forward 5′-ACTCCTACGGGAGGCAGCA-3′ and reverse 5′-GGACTACHVGGGTWTCTAAT-3′. The sample-specific 7 bp barcode was included in the primers used for multiplex sequencing. The PCR was performed in 25 μl of reaction mixture containing 5 μl buffer (5×), 0.25 μl of Fast pfu DNA polymerase (5 U/μl), 2 μl of dNTP (2.5 mM), 1 μl of each forward and reverse primer, 1 μl of cDNA, and 14.75 μl of ddH2O. The PCR reaction conditions were as follows: 98 °C for 5 min; 98 °C for 30 s; 53 °C for 30 s; 72 °C for 45 s and 72 °C for extension. This was repeated for 25 cycles and a final 72 °C for 5 min. The PCR products were excised from a 1.5% agarose gel and purified using Vazyme VAHTSTM DNA Clean Beads (Vazyme, Nanjing City, China). Then, the products were quantified using Quant-iT PicoGreen dsDNA detection kit (Invitrogen, Carlsbad, CA, USA) following the manufacturer’s suggested protocols.
Barcoded V3 and V4 amplicons were sequenced using the pair-end method by Illumina Miseq. Original pair-end sequences with a mean quality lower than 20, containing ambiguous bases, a sequence length shorter than 250 bp, chimeras, adaptor contamination or host contaminating reads were removed. The original pair-end sequence reads that passed our quality control criteria and contained a sequence overlap of at least 15 bp without any mismatch were assembled according to their overlapping sequences by software FLASH (Fast Length Adjustment of Short reads).
2.5. Taxonomic classification and annotation analysis
Sequences with ≥97% similarity were assigned to the same operational taxonomic units (OTUs). And each representative OUT was annotated taxonomic information using the UNOISE pipeline in USEARCH. Alpha diversity was applied to analyze the complexity of species diversity within groups, including the Observed-species, Chao1, ACE, Shannon, and Simpson indices. Beta diversity analysis was used to evaluate differences between groups using principal co-ordinates analysis (PCoA). All these indices were calculated using the quantitative insights into microbial ecology (QIIME) pipeline (Version 1.7.0) and displayed using R ‘WennDiagram’ package (Version 3.2.0). The taxonomic composition and abundance were visualized using software MEGAN and GraPhlAn, and the taxa abundances between samples or groups were statistically compared at the phylum, class, order, family, genus and species level, respectively. The PICRUSt was used to precalculated the predicted genes and their functions in this study.
3. Results
3.1. Annotation analysis
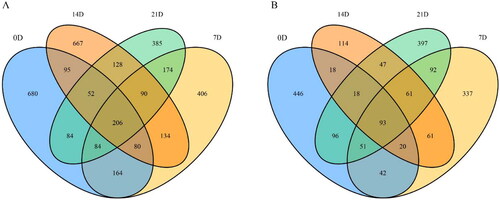
After high-throughput sequencing of 24 M. rosenbergii larvae and environmental water samples, a total of 911,722 OTUs were obtained. Among them, larvae received 514,871 OTUs, and environmental water received another 396,851. Venn diagram analysis identified 206 and 93 taxa were found to be shared between the larvae and environmental water among the stages, respectively (). The number of unique OTUs to the samples sampled by larvae on 0, 7, 14 and 21 days were 680, 406, 667 and 385, respectively. As well as they were 446, 337, 114 and 397 in environmental water on 0, 7, 14 and 21 days, respectively.
Figure 1. Common and specific OTU distribution of Macrobrachium rosenbergii larvae and environmental water at different stages. (A) OTU Venn diagram of Macrobrachium rosenbergii larvae at different stages. (B) OTU Venn diagram of environmental water at different stages. 1 D, 7 D, 14 D, and 21 D represent samples taken on 0th, 7th, 14th, and 21th days, respectively.
3.2. Microbial diversity of M. rosenbergii and environmental water at different stages
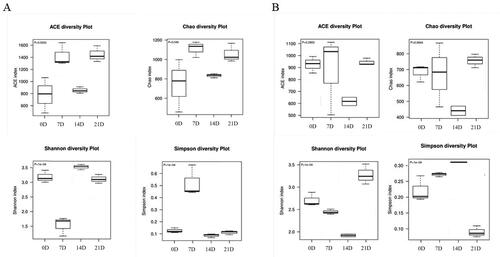
The richness and homogeneity of bacterial in larvae and environmental water samples were compared by Shannon, ACE Chao1, Simpson indexes in this study (). The ACE and Shannon indexes differed significantly among different stages in larvae (p < 0.01). With regard to the bacterial diversity in environmental water, the Shannon and Simpson values were significantly different at different stages (p < 0.01). This phenomenon indicated that the richness and homogeneity of samples from larvae and environmental water at different stages have undergone significant changes.
Figure 2. Alpha diversity analysis of OTUs. (A) Alpha diversity index of Macrobrachium rosenbergii larvae; (B) Alpha diversity index of environmental water.
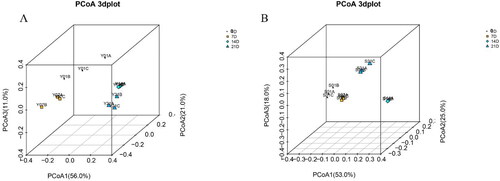
PCoA analysis using the weighted_UniFrac similarity metric revealed that the samples clustered according to age stages (). In the three-dimensional space composed of the principal coordinates PCoA1, PCoA2 and PCoA3, the spatial distances between larvae and environmental water at 0th, 7th, 14th and 21th days were significantly different, while the spatial distances of samples within the group were small. These results indicated that the bacterial community composition of the samples between the larvae and environmental water group were quite different, and the samples within the group had good repeatability.
3.3. Microbial composition of M. rosenbergii larvae and environmental water at different stages
As presented in , there were obvious differences in the microbiota composition of larvae and environmental water at different stages. At the phylum level, the majority of the sequences in larvae belonged to the Proteobacteria (33.47–91.97%), Planctomycetes (1.77–59.36%), and Firmicutes (0.26–30.65%). However, the proportion of these phyla throughout the stages was different. Bacteroidetes were mainly present at 14th day, and Firmicute were the major phyla at 21th day. For environmental water, the dominant flora were Proteobacteria (32.5–72.62%), Bacteroidetes (26.1–62.82%) and Actinobacteria (0.99–7.41%). The abundance of Bacteroidetes decreased significantly at 14th and 21th days.
Figure 4. Microbiota composition at the phylum and genus levels. (A) The microbiota composition at the phylum level of larvae between different stages. (B) The microbiota composition at the genus level of larvae between different stages. (C) The microbiota composition at the phylum level of environmental water between different stages. (D) The microbiota composition at the genus level of environmental water between different stages.
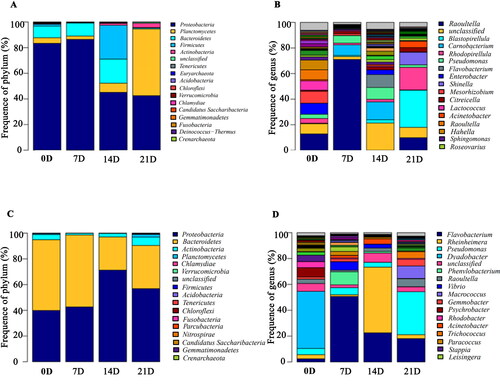
At the genus level, Raoultella (0.01–81.57%), Blastopirellula (0.37–34.86%), Carnobacterium (0.01–23.33%) and Rhodopirellula (0.97–19.96%) were found the dominant flora in larvae, while Flavobacterium (1.8–50.95%), Rheinheimera (1.11–50.62%), Pseudomonas (1.75–34.6%) and Dyadobacter (0.01–50.46%) were the dominant flora in environmental water. For larvae, Raoultella increased significantly at 7th day and decreased significantly at 14th day, and Blastopirellula increased significantly at 21th day. For environmental water, the abundance of Dyadobacter was significantly higher at 1th day than other stages. However, the abundance of Flavobacterium was significantly higher at 7th day than other stages. Additionally, Pseudomonas had the highest abundance at 21th day.
3.4. Kegg pathway analysis of M. rosenbergii larvae and environmental water at different stages
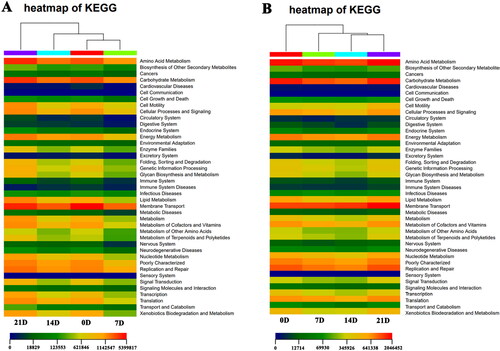
Based on the PICRUSt software, the differential flora in samples of larvae and environmental water at different stages were enriched into the KEGG pathway. And, the most obvious pathways for the enrichment of differential flora in samples at different periods were shown in . It was worth noting that several pathways enriched at different stages of larvae and environmental water were closely related to the breeding of larvae, such as Amino acid metabolism, Immune system, Immune system disease, Nervous system and Sensory system pathways.
4. Discussion
M. rosenbergii has always been considered a suitable species for aquaculture because it can grow in fresh water and low salt water. Because of its good growth performance and survival rate, M. rosenbergii has become a species promoted by the Food and Agriculture Organization of the United Nations (FAO) to the world, especially to developing countries. Nowadays, M. rosenbergii is widely cultivated throughout Southeast Asia, and plays an important role in aquaculture and fisheries (Sung et al. Citation2000; Lalrinsanga et al. Citation2014). M. rosenbergii is often interfered by many factors in the process of growth and development (Dong et al. Citation2019). One of the important factors is the microbial community structure of M. rosenbergii larvae and the environment water (Xie et al. Citation2017; Ni et al. Citation2019).
In our findings, the results of OTU analysis between M. rosenbergii larvae and environmental water showed that there were quite different at different stages. The alpha diversity analysis showed that the abundance and uniformity of the flora of larvae and water samples were significantly different at four stages. The beta diversity analysis also illustrated the differences in microbial community between larvae and environmental water during developmental stages. These results suggest that there may be some flora that affect the growth and development of larvae at different stages. It provides us with some clues to find some flora that affects the growth and development of M. rosenbergii.
The results of the taxonomic composition of species further showed the difference in flora of M. rosenbergii larvae and environmental water at different stages. The dominant flora at the phylum level of larvae is Proteobacteria, Plantomycetes, Firmicutes and Bacteroidetes, while at the genus level of genus is Raoultella, Blastopirellula, Carnobacterium and Rhodopirellula. However, the dominant flora of environmental water was obviously different from that of larvae. Proteobacteria, Bacteroidetes and Actinobacteria were the predominant flora at phylum level, while Flavobacterium, Rheinheimera, Pseudomonas and Dyadobacter were the predominant indexes at phylum level. These findings were basically consistent with those of Dong et al. (Citation2019). Among these different flora, the abundance of Raoultella in larvae increased significantly at 7th day and decreased significantly at 14th day. And the abundance of Blastopirellula increased significantly at 21th day. Raoultella is a human pathogenic bacteria, which has not yet been reported in shrimp (Appel et al. Citation2021), and its pathogenicity remains to be further studied. Blastopirellula is a kind of planktonic bacteria. The peptidoglycan-free cell wall of planktonic bacteria allows them to resist the action of several antibacterial compounds produced by macroalgae or other bacteria in the biofilm community. And these compounds can effectively resist biological contamination by other microorganisms (Lage and Bondoso Citation2014). Therefore, the abundance changes of Blastopirellula in different stages of larvae are also a direction that we need to study in the future. In environmental water, the abundance of Dyadobacter was highest at 1th day, as well as the abundance of Flavobacterium and Pseudomonas were highest at 7 days and 21 days, respectively. Flavobacterium is the pathogen of bacterial cold water disease, which can cause the death of rainbow trout (Li et al. Citation2021). However, the effect of Flavobacterium on M. rosenbergii larvae has not been reported yet. Pseudomonas plecoglossicida is a Gram-negative aerobic rod-shaped bacterium. It is the pathogen of visceral leukoplakia of cultured fish and can cause serious economic losses (Jiao et al. Citation2021). Therefore, both of Flavobacterium and Pseudomonas plecoglossicida are also the pathogens that we need to focus on during the breeding period of M. rosenbergii.
In order to explain the function of the differential flora in the larvae and environmental water, we predicted the pathway of the enrichment of the differential flora to the KEGG database based on the PICRUSt software. Among the obviously different pathways, Amino acid metabolism, Immune system, Immune system disease, Nervous system and Sensory system were closely related to the breeding of M. rosenbergii. Zhang et al. (Citation2021) compared the effects of different lysine isomers on the growth performance of M. rosenbergii, enzyme activities related to amino acid metabolism, antioxidant capacity and muscle amino acid composition. Their experimental results showed that M. rosenbergii can effectively use L-lysine for growth. D-Lysine can be used partially but shows specific toxic effects against oxidation. In our study, the Amino acid metabolism pathway of larvae has undergone significant changes at different stages. Combined with the results of Zhang et al., it was inferred that the metabolism of amino acids in the larvae had changed at different stages. Another study showed that Staphylococcus aureus and Vibrio parahaemolyticus infections can significantly up-regulate the total SOD activity and transcription level of M. rosenbergii MnSOD, thereby affecting the immune system (Li et al.Citation2021). Significant changes occurred in the pathways of Immune system and Immune system disease at different stages of M. rosenbergii larvae and water bodies, which means that there was a clear correlation between the flora of larvae and environmental water and the immune function of larvae. In addition, the KEGG pathway were also enriched in the Nervous system pathway. Neuropeptide is an endogenous active substance that exists in nerve tissue and participates in the function of the nervous system, and it plays a key role in controlling the feeding and reproduction of prawns (Tinikul et al.Citation2022). Neuropeptides integrate sensory stimuli including light, chemistry and taste reception to control feeding and reproduction, especially for ovarian development of female M. rosenbergii (Thongrod et al. Citation2019). The significant enrichment of the different flora in the Nervous system and Sensory system pathways means that these different flora may play a beneficial role in the development of M. rosenbergii in the Nervous system and Sensory system pathways.
5. Conclusion
In this study, there are significant changes in the flora of M. rosenbergii larvae and environmental water at different stages. The main changes in the flora of larvae at the genus level were Raoultella and Blastopirellula. In addition, the predicted pathways of amino acid metabolism, immune system, immune system disease, nervous system and sensory system were closely related to the breeding of M. rosenbergii. All in all, these discovered flora and metabolic pathways might provide theoretical guidance for the high-quality breeding of M. rosenbergii.
Ethics statement
The sample collection and experiments in the study were in compliance with the recommendations of the Laboratory Animals (Ministry of Science and Technology of China 2006) and approved by the Animal Ethics Committee of Zhejiang Institute of Freshwater Fisheries (Animal Ethics no. 1067, March 6, 2019).
Acknowledgement
None.
Disclosure statement
All authors declare that they have no conflict of interest.
Additional information
Funding
References
- Appel TM, Quijano-Martinez N, De La Cadena E, Mojica MF, Villegas MV. 2021. Microbiological and clinical aspects of Raoultella spp. Front Public Health. 9:686789.
- Dong XX, Lv HL, Zhao WY, Yu YB, Liu QG. 2019. Effects of different cultural patterns on microbial communities in the intestine of Macrobrachium rosenbergii and interactions with environment factors. J Shangh Ocean Univ (China). 28(04):501–510.
- Jiao J, Zhao L, Huang L, Qin Y, Su Y, Zheng W, Zhang J, Yan Q. 2021. The contributions of flig gene to the pathogenicity of Pseudomonas plecoglossicida and pathogen-host interactions with Epinephelus coioides. Fish Shellfish Immunol. 119:238–248.
- Lage OM, Bondoso J. 2014. Planctomycetes and macroalgae, A striking association. Front Microbiol. 5:267.
- Lalrinsanga PL, Pillai BR, Patra G, Mohanty S, Naik NK, Das RR, Sahu S, Nelliyoura R. 2014. Yield characteristics and morphometric relationships of giant freshwater prawn, Macrobrachium rosenbergii (de Man, 1879). Aquacult Int. 22(3):1053–1066.
- Laudadio I, Fulci V, Palone F, Stronati L, Cucchiara S, Carissimi C. 2018. Quantitative assessment of shotgun metagenomics and 16S rDNA amplicon sequencing in the study of human gut microbiome. OMICS. 22(4):248–254.
- Lee S, Suwa M, Shigemura H. 2019. Metagenomic analysis of infectious F-specific RNA bacteriophage strains in wastewater treatment and disinfection processes. Pathogens 8(4):217.
- Li P, Cai HY, Zhu Y. 2002. Application test of effective microorganisms in the culture of Macrobrachium rosenbergii. J Aquacult (China). (04):23–24.
- Li S, Chai J, Knupp C, Nicolas P, Wang D, Cao Y, Deng F, Chen F, Lu T, Loch TP. 2021. Phenotypic and genetic characterization of Flavobacterium psychrophilum recovered from diseased salmonids in China. Microbiol Spectr. 9(2):e0033021.
- Liu ZG, Lu MX, Ke XL, Wang S, Zhang DF. 2018. Correlation between microflora structure in intestinal tract and aquaculture environment of tilapia (Oreochromis niloticus) and Strptococcicosis. J Fish China. 42(10):13.
- Li Y, Zhan F, Li F, Lu Z, Shi F, Xu Z, Yang Y, Zhao L, Qin Z, Lin L. 2021. Immune function of cytosolic manganese superoxide dismutase from Macrobrachium rosenbergii in response to bacterial infection. Aquaculture 541:736771.
- Li X, Zhou Y, Jiang Q, Yang H, Pi D, Liu X, Gao X, Chen N, Zhang X. 2019. Virulence properties of Vibrio vulnificus isolated from diseased zoea of freshness shrimp Macrobrachium rosenbergii. Microb Pathog. 127:166–171.
- Madhavan A, Sindhu R, Parameswaran B, Sukumaran RK, Pandey A. 2017. Metagenome analysis: a powerful tool for enzyme bioprospecting. Appl Biochem Biotechnol. 183(2):636–651.
- Nguyen TMT, Chen TY, Shiau CY, Cheng YT, Chang YW. 2019. Study on biochemical divergences of the meat and egg of freshwater prawns (Macrobrachium rosenbergii). Food Sci Nutr. 7(6):2017–2023.
- Ni M, Gao Q, Yuan JL, Chen XF. 2019. Effect of saiinity on the water quality and microbiai community structure of the water for young Macrobrachium rosenbergii rearing. Acta Agricult Universi Jiangx (China). 41(05):976–985.
- Potapov SA, Tikhonova IV, Krasnopeev AY, Kabilov MR, Tupikin AE, Chebunina NS, Zhuchenko NA, Belykh OI. 2019. Metagenomic analysis of virioplankton from the pelagic zone of Lake Baikal. Viruses. 11(11):991.
- Qin Y. 2001. Application of EM in the nursery of Macrobrachium rosenbergii. J Guangxi Fish Sci Technol (China). 2(2):37–38.
- Shang LQ, Xue SK, Wang XJ, Zhang XL. 2019. Metagenomic comparative study on the intestinal microbial diversity of stocked cloven-hoofed animals in high elevation area of Northwest China. J Anhui Agricult Sci (China). 47(07):98–101.
- Sudarikov K, Tyakht A, Alexeev D. 2017. Methods for the metagenomic data visualization and analysis. Curr Issues Mol Biol. 24:37–58.
- Sung HH, Hwang SF, Tasi FM. 2000. Responses of giant freshwater prawn (Macrobrachium rosenbergii) to challenge by two strains of Aeromonas spp. J Invertebr Pathol. 76(4):278–284.
- Thongrod S, Wanichanon C, Sobhon P. 2019. Distribution of neuropeptide F in the ventral nerve cord and its possible role on testicular development and germ cell proliferation in the giant freshwater prawn, Macrobrachium rosenbergii. Cell Tissue Res. 376(3):471–484.
- Tinikul Y, Kruangkum T, Tinikul R, Sobhon P. 2022. Comparative neuroanatomical distribution and expression levels of neuropeptide f in the central nervous system of the female freshwater prawn, Macrobrachium rosenbergii, during the ovarian cycle. J Comparat Neurol. 530(4):729–755.
- Wang GL, Liu JJ. 2017. Macrobrachium rosenbergii breeding and attention issues. Sci Fish Farm (China). (12):87.
- Xie Q, Jiang M, Hu CF, Zhang JL, Wang CF, Tong XL, Gu DP, Hu WG, Yu ZL, Dai XL. 2017. Microbial community diversity on different substrates in Macrobrachium rosenbergii over-wintering cultivation. Microbiol China. 44(02):336–347.
- Yang G, Frinsko M, Chen X, Wang J, Hu G, Gao Q. 2012. Current status of the giant freshwater prawn (Macrobrachium rosenbergii) industry in China, with special reference to live transportation. Aquac Res. 43(7):1049–1055.
- Zhang Y, Ma XS, Jing RY, Ma FY, Guo JY, Wang YP, Wang HT. 2019. Effects of successive-planting poplar plantation on soil microbial community. J Shand Univ (Nat Sci China). 54(01):36–46.
- Zhang X, Wang H, Zhang J, Lin B, Chen L, Wang Q, Li G, Deng J. 2021. Utilization of different lysine isomers: a case study on the growth, metabolic enzymes, antioxidant capacity and muscle amino acid composition in Macrobrachium rosenbergii. Anim Feed Sci Technol. 280:115078.
- Zhu H, Xiao C, Shang HQ, Guo XY, Du XP, Qin C. 2019. Analysis of gut microbiomes of Rhesus macaques of different ages by high-throughput sequencing. Acta Laborat Anim Sci Sin (China). 27(01):72–78.