Abstract
Objective
PM2.5 is closed linked to asthma exacerbation. The Notch1 pathway acts as an important pathway, ultimately inducing T-helper cells that express GATA3 and its corresponding Th2 cytokines. The regulatory effects of miR-139-5p on the Notch1 pathway have been indicated in cancer. However, studies on miR-139-5p have not applied asthma-related models. The role of miR-139-5p and its regulatory effects on the Notch1-GATA3 pathway in asthma exacerbation induced by acute PM2.5 exposure has not been elucidated. We hypothesize that acute PM2.5 exposure induces asthma exacerbation by regulating the expression of miR-139-5p and activating the Notch1-GATA3 pathway.
Methods
We first employed Diseased Human Bronchial Epithelial Cells-Asthma cells to establish an in vitro model of acute exposure to PM2.5, and explored the relationship between the different concentrations and durations of acute PM2.5 exposure and the activation of Notch1-GATA3 pathway. We investigated the protein and mRNA expression changes of Notch1, upstream Jagged1, downstream GATA3, as well as the regulatory effect of miR-139-5p involved in it.
Results
The miR-139-5p expression increased within 24 h of PM2.5 exposure. However, if PM2.5 exposure was sustained, miR-139-5p expression turned to decrease, accompanied by upregulations of the mRNA and protein expression of Notch1-GATA3 pathway. Overexpression of miR-139-5p blocked Notch1-GATA3 pathway activation induced by acute PM2.5 exposure.
Conclusion
Acute PM2.5 exposure can activate Notch1-GATA3 pathway in asthma bronchial epithelial cells model, which might be involved in PM2.5-induced asthma exacerbation. miR-139-5p has a potential protective role of inhibiting PM2.5-induced asthma airway inflammation by targeting Notch1.
Keywords:
Introduction
Asthma is a common chronic airway inflammatory disease impacting more than 300 million people globally (Citation1). Asthma exacerbation accelerates disease progression and increases the incidence of hospitalization and death; in addition, it contributes to functional loss and increased healthcare costs (Citation2). Therefore, reducing the risk of asthma exacerbation is relevant both clinically and socially. At present, it is believed that outdoor air pollution plays an important role in asthma exacerbations (Citation3). As one of the essential components of outdoor air pollutants, fine particulate matter (PM2.5) which is classified as particles with an aerodynamic diameter ≤ 2.5 μm, is being intensely studied for its effects on the human body. Hopke’s research has demonstrated that higher concentration PM2.5 exposure (with an interquartile interval of 6.8 μg/m3) was associated with 0.3%∼0.9% increase risk of outpatient visits for asthma exacerbation (Citation4). An investigation revealed there was an 0.65% increase risk of asthma exacerbation related hospitalizations and an 0.65% increase risk of asthma exacerbation related emergency visits for every 10 μg/m3 increase in PM2.5 concentration (Citation5). Our systematic review and meta-analysis revealed that exposure to outdoor air pollution increases the risk of asthma exacerbation on the same day, with an increase of 10 μg/m3 in PM2.5 concentration resulting in a 0.7% increase risk of asthma exacerbation (Citation6).
When severe air pollution occurs, in addition to advising asthma patients to limit outdoors activity, use air purification devices, and wear masks when venturing outside, other intervention methods are being explored to prevent asthma acute exacerbations. Our prospective randomized controlled trial proposed a novel rescue intervention strategy of inhaling corticosteroids within 24 h of severe outdoor air pollution exposure, and the results showed that this strategy could effectively reduce the risk of asthma exacerbation in the intervention group (Citation7). However, the mechanisms by which acute PM2.5 exposure induces asthma exacerbation remain unclear, and effective interventional targets still need to be further explored.
Our previous research found that the expression of numerous microRNAs (miRNAs) was significantly changed after exposure to PM2.5 in healthy mouse models or in normal human bronchial epithelial cells. MicroRNA-139-5p (miR-139-5p) was the miRNA that underwent the earliest and most significant changes after exposure to PM2.5. The research demonstrated that miR-139-5p expression of mouse lung tissue increased after one day of PM2.5 exposure, however, the miR-139-5p expression turned to decrease after seven days of PM2.5 exposure (Citation8). We also demonstrated that miR-139-5p was capable of targeting Notch1 mRNA, and overexpression miR-139-5p could suppress Notch1 protein expression in normal human bronchial epithelial cells. The results revealed that after chronic PM2.5 exposure, Notch1 protein expression significantly increased, while miR-139-5p expression decreased (Citation9). The Notch1 pathway, including the Notch1 ligand Jagged1, from the dendritic cells or antigen-presenting cells acts as an important pathway, ultimately inducing T-helper cells that express GATA3 and its corresponding Th2 cytokines, IL-4, IL-5, and IL-13 (Citation10). The regulatory effects of miR-139-5p on the Notch1 pathway have been indicated in cancer cells (Citation11). However, these previous studies on miR-139-5p have not applied asthma related models. And the expression changes of miR-139-5p showed a opposite trend with different PM2.5 exposure durations. Therefore, the role of miR-139-5p and its regulatory effects on the Notch1-GATA3 pathway in asthma exacerbation induced by acute PM2.5 exposure has not been elucidated.
In this study, we first employed Diseased Human Bronchial Epithelial Cells-Asthma (D-HBE-As) cells to establish an in vitro model of acute exposure to PM2.5, and explored the relationship between the different concentrations and durations of acute PM2.5 exposure and the activation of Notch1-GATA3 pathway. We investigated the protein and mRNA expression changes of Notch1, its upstream ligand Jagged1, and downstream GATA3, as well as the regulatory effect of miR-139-5p involved in it. We hypothesize that acute PM2.5 exposure induces asthma exacerbation by regulating the expression of miR-139-5p and activating the Notch1-GATA3 pathway.
Methods and materials
PM2.5 collection and preparation
The PM2.5 was collected by high volume sampler system (Staplex PM2.5 SSI, USA), which was set on the top building of Peking University First Hospital located in the central area of Beijing. The collection method and component analysis report of PM2.5 had been described in previous published literature (Citation8). The PM2.5 suspension was obtained by thoroughly mixing the particulate matter and culture medium with 1% fetal bovine serum under sonication to stimulate cells.
Cell culture and treatment
The Diseased Human Bronchial Epithelial Cells-Asthma (D-HBE-As, MeisenCTCC, China) cells were cultured in Roswell Park Memorial Institute 1640 (RPMI1640) with L-Glutamine (VivaCell, China) supplemented with 10% fetal bovine serum (VivaCell, China), 100 U/ml penicillin and 100 μg/ml streptomycin (VivaCell, China) and maintained at 37 °C in a saturated humidity atmosphere containing 5%CO2. D-HBE-As cells were treated with different concentrations of PM2.5 (low dose = 50 μg/ml, middle dose = 100 μg/ml, high dose = 200 μg/ml) for different hours (0, 6, 12, 24, 48 h) to mimic acute exposure. For gain-of-function, miR-139-5p mimic was used to induce an elevation in the levels of miR-139-5p within cells. For loss-of-function, miR-139-5p inhibitor was used to diminish gene silencing effect by selectively inhibiting miR-139-5p. Their negative controls were mimic negative control and inhibitor negative control. miR-139-5p mimic, mimic negative control, miR-139-5p inhibitor and inhibitor negative control (RIBOBIO, China) were complexed respectively with LabFect RNAi Transfection Kit (LABLEAD, China) according to the manufacturer’s instructions. D-HBE-As cells were transfected respectively with miR-139-5p mimic (50 nM), miR-139-5p inhibitor (50 nM) and their negative controls (50 nM) for 72h followed by PM2.5 (200 μg/ml) for different hours (0, 6, 12, 24, 48 h).
Western blot for protein
The proteins expressions were detected by western blotting. The total proteins were extracted with a precooling RIPA lysis buffer containing 1% phosphatase inhibitor cocktail and 1% proteinase inhibitor cocktail (LABLEAD, China). The cell lysate was centrifuged at 12,000 rpm for 15 min at 4 °C. Subsequently, the supernatant fluid was collected. The protein content was determined using the PierceTM Rapid Gold BCA Protein Assay Kit (Thermo Scientific, USA) according to the manufacturer’s instructions. The proteins were separated through sodium dodecyl sulfate-polyacrylamide gel electrophoresis. The protein samples were transferred to PVDF membranes and blocked with 5% nonfat milk. The transferred membranes were incubated with Jagged1 (1:500, Abcam, USA), Notch1 (1:500, Abcam, USA), GATA3 (1:500, Abcam, USA) and β-actin (1:2500, LABLEAD, China) antibody overnight on a shaker at 4 °C, followed by the appropriate secondary antibody (1:5000, LABLEAD, China) for 1h at room temperature. The proteins were quantified and visualized using the Syngene GeneGenius (GBOX-CHEMI-XT4, SYNGENE, USA).
Quantitative assessment of mRNA and miRNA expression
The RNA Easy Fast Tissue/Cell Kit (TIANGEN, China) was used to extract total RNA. The RNA from each sample was quantified by Nanodrop 2000 (Thermo Fisher Scientific, USA). For mRNA, 500 ng total RNA was reversely transcribed to 10 μl complementary DNA (cDNA) by using PrimeScriptTM RT Master Mix Kit (TAKARA, Japan). For microRNA (miRNA), 750 ng total RNA was reversely transcribed to 10 μl cDNA by using Mir-XTM miRNA First-Stand Synthesis Kit (TAKARA, Japan). The specific primers of mRNA and miRNA (TIANYIHUIYUAN, China) used for qRT-PCR were shown in . The mRQ 3’primer, U6 forward primer and U6 reverse primer were included in the Mir-XTM miRNA First-Stand Synthesis Kit (TAKARA, Japan). qRT-PCR was carried out in a 20 μl reaction system containing 2 μl cDNA and 10 μl TB Green Premix Ex TaqTM II (TAKARA, Japan) on Step One Plus Real-Time PCR System (7500, Applied Biosystems, USA). The relative expressions of mRNA or miRNA were calculated by 2-△△CT method and normalized to the expressions of β-action or U6 respectively.
Table 1. The specific primers used for qRT-PCR.
Statistical analysis
Statistical analysis was performed with SPSS for Windows, version 22.0 (SPSS Inc., USA). Statistical differences between experimental groups and control groups were evaluated with Independent Samples T-Test. The experimental data were homogeneity of variance by Levene’s Test. The results of ‘equal variances assumed’ line were used for the data of homogeneity of variance. The results of ‘equal variances not assumed’ line were used for the data of homogeneity of variance. All tests were two-tailed. Statistical significance was defined at a level of 5% (p < 0.050). Difference significance among groups was assessed as * p < 0.050, ** p < 0.010, *** p < 0.000.
Results
Acute PM2.5 exposure activated Notch1-GATA3 signal pathway of D-HBE-As cells
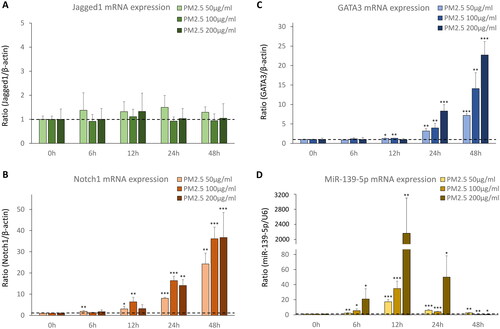
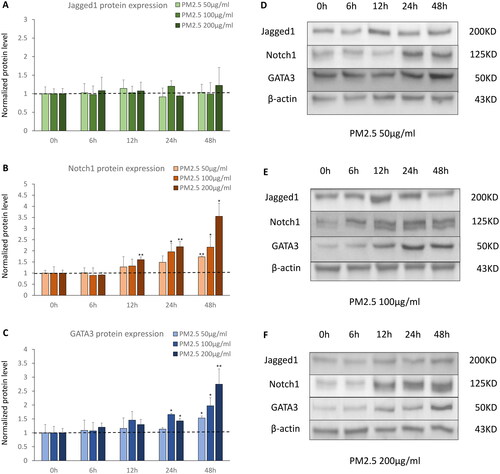
We treated D-HBE-As cells with different concentrations of PM2.5 (low dose = 50 μg/ml, middle dose = 100 μg/ml, high dose = 200 μg/ml) for different hours (0, 6, 12, 24, 48 h) to mimic acute PM2.5 exposure. We found Jagged1 protein expression remained unaffected by varying concentrations and durations of PM2.5 exposure (). However, acute PM2.5 exposure upregulated the protein expression levels of Notch1 and GATA3. Notch1 protein expression increased from as early as 12 h of PM2.5 exposure, and augmented further with the prolongation of PM2.5 exposure time. This prominent increase was even more noticeable and occurred earlier under higher concentrations of PM2.5 exposure (). GATA3 protein expression increased in 24 h being exposed to PM2.5, and the timing was slightly later than that of Notch1.The trend of GATA3 protein expression in response to PM2.5 concentrations and exposure durations was analogous to that of Notch1 ().
Figure 1. The protein expression of Notch1-GATA3 pathway during acute PM2.5 exposure (n = 3). (A, B and C) The normalized protein levels of Nothc1-GATA3 pathway in D-HBE-As cells treated with different concentrations of PM2.5 (50, 100, 200 μg/ml) for different hours (0, 6, 12, 24, 48 h) to mimic acute exposure. (D, E and F) The Western Blot results of Nothc1-GATA3 pathway in D-HBE-As cells during acute PM2.5 exposure. *p < 0.050, **p < 0.010, ***p < 0.000, compared with 0h group as control.
The mRNA expression of miR-139-5p and its potential target Notch1 during acute PM2.5 exposure
To observe miR-139-5p expression and its potential target Notch1 in PM2.5 treated D-HBE-As cells, we detected the mRNA expression levels of miR-139-5p and Notch1-GATA3 pathway by qRT-PCR. Jagged1 mRNA expression remained unchanged in response to different concentrations of PM2.5 and exposure durations (). Notch1 mRNA expression increased as early as 6 h of exposure. And GATA3 mRNA expression significantly increased at 12 h of exposure, lagging behind Notch1. Both levels of Notch1 and GATA3 mRNA expression continued increasing with longer exposure time and higher concentration of PM2.5 (). After PM2.5 exposure, miR-139-5p expression started to increase significantly at 6 h and reached its peak at 12 h. This increasing trend was more pronounced with the rise of PM2.5 concentration. Though the levels of miR-139-5p expression were still above that of pre-exposure at 24 h exposure, the peak value seen at 12 h had decreased. At 48 h exposure, miR-139-5p expression decreased further, and the decrease became more pronounced with increase of PM2.5 concentration. In the higher dose groups of PM2.5 (100 μ g/ml, 200 μ g/ml), the levels of miR-139-5p expression were significantly lower than that of pre-exposure ().
Inhibiting miR-139-5p expression upregulated the protein expression levels of Notch1 and GATA3 in absent of PM2.5
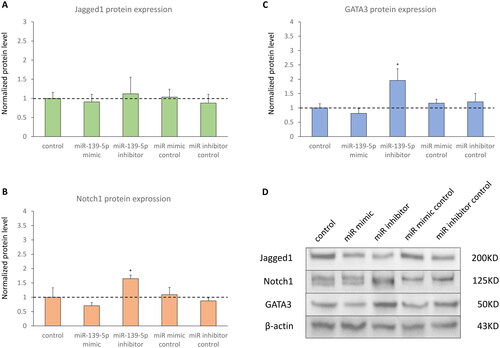
D-HBE-As cells were transfected with miR-139-5p mimic, miR-139-5p inhibitor, or scrambled controls to examine the effect of miR-139-5p on its potential target Notch1. The efficiency of transfection was shown in . No significant alterations in mRNA and protein expression levels of Jagged1 were observed in transfected cells ( and ). Notch1 protein expression significantly increased in miR-139-5p inhibitor transfected cells, while Notch1 mRNA expression was not significantly different among all groups (). Inhibiting miR-139-5p expression led to a notable increase in both mRNA and protein expression of GATA3 ( and ). In absent of PM2.5 exposure, overexpression of miR-139-5p did not seem to have a significant impact on the Notch1 pathway in D-HBE-As cells, except for upregulating the expression of GATA3 mRNA.
Figure 3. The mRNA expression of Notch1-GATA3 pathway and miR-139-5p in D-HBE-As cells transfected with miR-139-5p mimic, inhibitor, or scrambled controls (n = 4). *p < 0.050, **p < 0.010, ***p < 0.000, compared with control group.
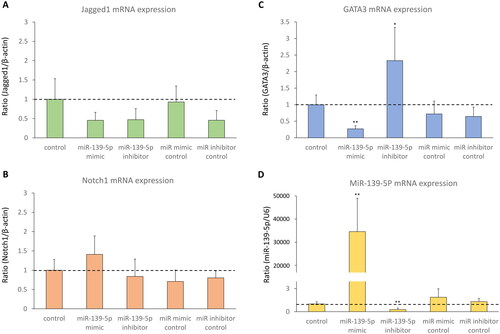
Figure 4. The protein expression of Notch1-GATA3 pathway in D-HBE-As cells transfected with miR-139-5p mimic, inhibitor, or scrambled controls (n = 3). (A, B and C) The normalized protein levels of Nothc1-GATA3 pathway. (D) The Western Blot results of Nothc1-GATA3 pathway. *p < 0.050, **p < 0.010, ***p < 0.000, compared with control group.
The negative regulatory effect of miR-139-5p on the expression of Notch1 and GATA3 in acute PM2.5 exposure
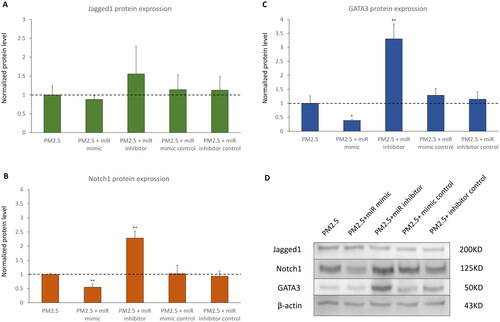
To discover whether miR-139-5p could regulate the expression of Notch1 and GATA3 in acute PM2.5 exposure treated D-HBE-As cells, cells were transfected with miR-139-5p mimic, miR-139-5p inhibitor and scrambled controls following exposure to PM2.5 (200 μg/ml) for 48 h. The efficiency of transfection was shown in , while the magnitude of miR-139-5p mRNA increase after mimic transfection was less in cells exposed to PM2.5 than in cells without PM2.5 exposure ( and ). Following exposure to PM2.5, Jagged1 displayed no substantial alterations in its mRNA and protein expression levels in transfected cells ( and ). The expression of Notch1 mRNA did not change in miR-139-5p mimic and inhibitor transfected cells (Figure5B). However, overexpression of miR-139-5p downregulated Notch1 protein expression, and inhibiting the expression of miR-139-5p had the opposite effect of upregulating Notch1 protein expression (). It suggested that the regulatory role of miR-139-5p in Notch1 pathway was to inhibit the transcription of Notch1 mRNA into protein. Overexpression of miR-139-5p also downregulated the expression levels of GATA3 mRNA and protein, while miR inhibitor had the opposite effect ( and ).
Figure 5. The mRNA expression of Notch1-GATA3 pathway and miR-139-5p during acute PM2.5 exposure in D-HBE-As cells transfected with miR-139-5p mimic, inhibitor, or scrambled controls (n = 4). *p < 0.050, **p < 0.010, ***p < 0.000, compared with only PM2.5 exposure group as control.
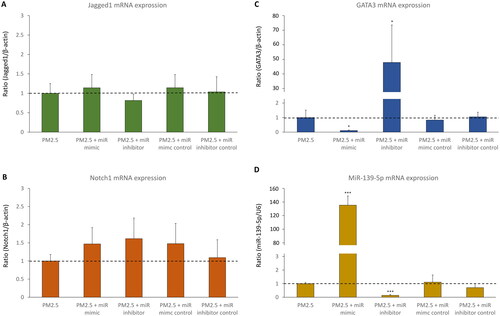
Figure 6. The protein expression of Notch1-GATA3 pathway during acute PM2.5 exposure in D-HBE-As cells transfected with miR-139-5p mimic, inhibitor, or scrambled controls (n = 3). (A, B and C) The normalized protein levels of Nothc1-GATA3 pathway. (D) The Western Blot results of Nothc1-GATA3 pathway. *p < 0.050, **p < 0.010, ***p < 0.000, compared with only PM2.5 exposure group as control.
Discussion
To date, numerous studies have indicated that acute exposure of PM2.5 can increase airway responsiveness and induce acute exacerbation of asthma (Citation12–14). However, the exact mechanism of PM2.5 causing aggravation of asthma remains unclear. Our study showed that acute PM2.5 exposure activated Notch1-GATA3 pathway in D-HBE-As cells. We found that Notch1 mRNA and protein expression in D-HBE-As cells increased as early as 6 h and 12 h of PM2.5 exposure respectively. After a slight delay, GATA3 mRNA and protein expression also began to increase from 12 h and 24 h being exposed to PM2.5 respectively. The expressions of Notch1 and GATA3 in D-HBE-As cells were both significantly enhanced with the increase of PM2.5 exposure concentration and duration. We further discovered that miR-139-5p played an important role in the regulation of Notch1-GATA3 signaling pathway. miR-139-5p negatively regulated the expression of Notch1 and GATA3 proteins during acute PM2.5 exposure, which was demonstrated by transfecting D-HBE-As cells with mimic or inhibitor of miR-139-5p. The Notch signaling pathway is a highly conserved signal transduction pathway. There are four homologous receptors (Notch1-4) in mammals, which are bound by five ligands (Delta-like 1,3, 4, and Jagged1, 2) (Citation15). The Notch receptor is a single transmembrane receptor protein. Through intercellular interaction and ligand binding, as well as through enzymatic hydrolysis by γ-secretase, Notch activates the downstream target genes of the Hes and Hey family, which together participate in the activation, proliferation, and differentiation of T cells (Citation16,Citation17). Notch1 has a significant role in the differentiation of Th1, Th2, Th17, and Treg cells (Citation18–21). In murine model of asthma, raised expression of Notch1 receptor following OVA challenge was observed (Citation22,Citation23). Blocking of Notch1 signaling by Notch1-specific small interfering RNA or by γ-secretase inhibitor, an effective blocker of Notch1, significantly ameliorated the airway inflammation infiltration, accompanied with the decreasing of IL-4 and increasing of IFN-γ (Citation24,Citation25). In children with allergic asthma, the upregulation of Notch1 mRNA expression in peripheral blood mononuclear cells was also observed (Citation26).
GATA3 as a direct transcriptional Notch target acts in concert with Notch signaling to generate optimal Th2 cell responses (Citation27). The introduction of an activated allele of Notch1 into CD4 + T cells led to the specific and direct upregulation of a developmentally regulated Gata3 transcript, which acts in concert with Notch signaling to synergistically activate the IL-4 expression and the Th2 cell responses (Citation28). A significant increase in GATA3 expression was demonstrated in asthmatic airways compared with that in control subjects (Citation29,Citation30). In a transgenic murine model of asthma, all key features of asthma were severely attenuated by inhibiting the expression of GATA3 (Citation31). Taken together, Notch1–GATA3 signaling pathway plays an essential role in the control of production of Th2 cytokines, thereby influencing the onset and exacerbation of asthma.
However, the role of Notch1-GATA3 signaling pathway in the PM2.5 exacerbated asthma airway inflammation is less studied. Liu and colleagues demonstrated the important role of Notch signaling pathway in Muc5ac secretion induced by PM2.5 in Beas-2B cells and in rat asthma model (Citation32,Citation33). Wu and colleagues revealed that PM2.5 promoted the expression of the mRNA and protein of Notch1 and aggravated Th1/Th2 immune imbalance in asthmatic mice, while γ-secretase inhibitor partially inhibited the activation of the Notch signaling pathway and alleviated the aggravation of immune imbalance (Citation34). In a PM-exacerbated asthmatic mice model, PM exposure led to upregulation of GATA3 mRNA accompanied by excessive Th2 response (Citation35). Our present study confirmed that Notch1-GATA3 expression was upregulated in asthma human bronchial epithelial cells by acute exposure of PM2.5 in a time-dependent and dose-dependent way. We speculate that PM2.5 exposure may aggravate asthma airway inflammation by activating Notch1-GATA3 signaling pathway.
It has been reported by many studies that PM2.5 exposure can significantly alter the expression of various miRNAs, which are closely related with the inflammatory response induced by PM2.5 (Citation36). In a PM2.5 exposed mice model, our research team revealed that the levels of miR-139-5p in lung tissue increased significantly and reached its peak on the first day after PM2.5 exposure, and returned to the baseline level on the 7th day after exposure. The negative linear correlation between miR-139-5p and the IL-4/IFN-γ level of broncho-alveolar lavage fluid (BALF) was also observed (Citation8). We used luciferase analysis to identify the binding site and the interaction of miR-139-5p and Notch1, and confirmed that miR-139-5p directly targeted Notch1 (Citation9). In the chronic PM2.5 exposure murine model, we found PM2.5 induced downregulation of miR-139-5p expression and activated Notch1 pathway (Citation9).
The present study showed that the mRNA expression of miR-139-5p in D-HBE-As cells was significantly upregulated within a few hours after being exposed to PM2.5, peaked at 12h of exposure, and then downregulated thereafter. The result is consistent with the findings of our previous studies on acute exposure to PM2.5. We speculate that the transient increase in miR-139-5p expression after PM2.5 exposure might be a protective response. However, the upregulation of miR-139-5p cannot be maintained. When the D-HBE-As cells was exposed to high dose of PM2.5 (100 μ g/ml, 200 μ g/ml) for 48h, the expression of miR-139-5p was extremely inhibited, accompanied with activation of Notch1-GATA3 signaling pathway. The role of miR-139-5p on Notch1-GATA3 pathway was further investigated by using miR-139-5p mimic and inhibitor. When miR-139-5p was inhibited, both Notch1 and GATA3 were upregulated at protein levels in the absence of PM2.5 exposure. When the D-HBE-As was treated with PM2.5 (200 μg/ml) for 48 h, miR-139-5p exhibited a more noticeable regulatory role on Notch1-GATA3 pathway. Overexpression of miR-139-5p downregulated the expression of Notch1 and GATA3, while inhibiting miR-139-5p had the opposite effect. Our study confirmed that miR-139-5p played a negatively regulatory role on Notch1-GATA3 pathway in the acute PM2.5-exposed D-HBE-As cells. It was reported that miR-139-5p regulates diverse biological processes, and previous researches on its function have mainly focused on the field of cancer (Citation37). This is the first study to elucidate the regulatory role of miR-139-5p in Notch1 pathway activation induced by acute exposure to PM2.5, suggesting its potential role in asthma treatment.
However, our present study has some limitations. Although it was shown that miR-139-5p regulated the Notch1 pathway in bronchial epithelial cells of asthma patents after PM2.5 exposure, the physiological changes characteristic of asthma, such as airway hyperresponsiveness and Th2 airway inflammation, has not been investigated in this study. Moreover, several targets of miR-139-5p have been previously described (Citation37), such as FoxO1, Rho-kinase2, CXCR4, RAP1B, Type I Insulin-like GF, in addition to modulating Notch1-GATA3 signaling. Therefore, the results should be further validated using in vivo models.
Conclusion
Our results revealed that acute PM2.5 exposure activated Notch1-GATA3 pathway in asthma bronchial epithelial cells in vitro, which might be involved in the acute exacerbation of asthma. miR-139-5p played a protective role by targeting Notch1 and might serve as a potential intervention target in the future.
Authors’ contribution
Junjun Huang: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Resources, Validation, Visualization, Writing – original draft, Writing – review & editing. Yan Hu: Data curation, Formal analysis, Funding acquisition, Project administration, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing. Yunxia Wang: Methodology, Resources, Writing – original draft. Zhou Jin: Investigation, Resources, Writing – original draft.
| Abbreviation | ||
| BALF | = | broncho-alveolar lavage fluid |
| cDNA | = | complementary DNA |
| D-HBE-As | = | diseased human bronchial epithelial cells-asthma |
| miRNA | = | microRNA |
| miR-139-5p | = | microRNA-139-5p |
| PM2.5 | = | fine particulate matter |
| RPMI1640 | = | Roswell Park Memorial Institute 1640 |
Acknowledgements
We acknowledge the contribution of Dr. Xiaoyu Ma for consultation on cell culture technology.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of this article.
Additional information
Funding
References
- Rutter C, Silverwood R, Pérez Fernández V, Pearce N, Strachan D, Mortimer K, Lesosky M, Asher I, Ellwood P, Chiang CY, et al. The global burden of asthma. Int J Tuberc Lung Dis. 2022;26(1):20–23. PMID: 36284412.
- Huang K, Yang T, Xu J, Yang L, Zhao J, Zhang X, Bai C, Kang J, Ran P, Shen H, et al. Prevalence, risk factors, and management of asthma in China: a national cross-sectional study. Lancet. 2019;394(10196):407–418. doi:10.1016/S0140-6736(19)31147-X.
- Maio S, Sarno G, Tagliaferro S, Pirona F, Stanisci I, Baldacci S, Viegi G. Outdoor air pollution and respiratory health. Int J Tuberc Lung Dis. 2023;27(1):7–12. doi:10.5588/ijtld.22.0249.
- Hopke PK, Croft D, Zhang W, Lin S, Masiol M, Squizzato S, Thurston SW, van Wijngaarden E, Utell MJ, Rich DQ. Changes in the acute response of respiratory diseases to PM2.5 in New York State from 2005 to 2016. Sci Total Environ. 2019;677:328–339. doi:10.1016/j.scitotenv.2019.04.357.
- Tian Y, Xiang X, Juan J, Sun K, Song J, Cao Y, Hu Y. Fine particulate air pollution and hospital visits for asthma in Beijing, China. Environ Pollut. 2017;230:227–233. doi:10.1016/j.envpol.2017.06.029.
- Huang J, Yang X, Fan F, Hu Y, Wang X, Zhu S, Ren G, Wang G. Outdoor air pollution and the risk of asthma exacerbations in single lag0 and lag1 exposure patterns: a systematic review and meta-analysis. J Asthma. 2022;59(11):2322–2339. doi:10.1080/02770903.2021.2008429.
- Yang X, Huang J, Hu Y, Zhu S, Guo C, Wang X, Yang Z, Tian Z, Wang G. The rescue intervention strategy for asthma patients under severe air pollution: a single-center prospective randomized controlled trial. J Asthma. 2022;59(9):1712–1721. doi:10.1080/02770903.2021.1980584.
- Hou T, Liao J, Zhang C, Sun C, Li X, Wang G. Elevated expression of miR-146, miR-139 and miR-340 involved in regulating Th1/Th2 balance with acute exposure of fine particulate matter in mice. Int Immunopharmacol. 2018;54:68–77. doi:10.1016/j.intimp.2017.10.003.
- Wang Y, Zhong Y, Zhang C, Liao J, Wang G. PM2.5 downregulates MicroRNA-139-5p and induces EMT in bronchiolar epithelium cells by targeting Notch1. J Cancer. 2020;11(19):5758–5767. doi:10.7150/jca.46976.
- Tindemans I, van Schoonhoven A, KleinJan A, de Bruijn MJ, Lukkes M, van Nimwegen M, van den Branden A, Bergen IM, Corneth OB, van IJcken WF, et al. Notch signaling licenses allergic airway inflammation by promoting Th2 cell lymph node egress. J Clin Invest. 2020;130(7):3576–3591. doi:10.1172/JCI128310.
- Sun Q, Weng D, Li K, Li S, Bai X, Fang C, Luo D, Wu P, Chen G, Wei J. MicroRNA-139-5P inhibits human prostate cancer cell proliferation by targeting Notch1. Oncol Lett. 2018;16(1):793–800. doi:10.3892/ol.2018.8773.
- Lu X, Li R, Yan X. Airway hyperresponsiveness development and the toxicity of PM2.5. Environ Sci Pollut Res Int. 2021;28(6):6374–6391. doi:10.1007/s11356-020-12051-w.
- Luo J, Liu H, Hua S, Song L. The correlation of PM2.5 exposure with acute attack and steroid sensitivity in asthma. Biomed Res Int. 2022;2022:2756147–2756148. doi:10.1155/2022/2756147.
- Qibin L, Yacan L, Minli J, Meixi Z, Chengye L, Yuping L, Chang C. The impact of PM2.5 on lung function in adults with asthma. Int J Tuberc Lung Dis. 2020;24(6):570–576. doi:10.5588/ijtld.19.0394.
- Radtke F, Fasnacht N, Macdonald HR. Notch signaling in the immune system. Immunity. 2010;32(1):14–27. doi:10.1016/j.immuni.2010.01.004.
- Kopan R, Ilagan MX. The canonical notch signaling pathway: unfolding the activation mechanism. Cell. 2009;137(2):216–233. doi:10.1016/j.cell.2009.03.045.
- Zhou B, Lin W, Long Y, Yang Y, Zhang H, Wu K, Chu Q. Notch signaling pathway: architecture, disease, and therapeutics. Signal Transduct Target Ther. 2022;7(1):95. doi:10.1038/s41392-022-00934-y.
- Golub R. The Notch signaling pathway involvement in innate lymphoid cell biology. Biomed J. 2021;44(2):133–143. doi:10.1016/j.bj.2020.12.004.
- Laky K, Fowlkes BJ. Notch signaling in CD4 and CD8 T cell development. Curr Opin Immunol. 2008;20(2):197–202. doi:10.1016/j.coi.2008.03.004.
- Shang Y, Smith S, Hu X. Role of Notch signaling in regulating innate immunity and inflammation in health and disease. Protein Cell. 2016;7(3):159–174. doi:10.1007/s13238-016-0250-0.
- Zong D, Ouyang R, Li J, Chen Y, Chen P. Notch signaling in lung diseases: focus on Notch1 and Notch3. Ther Adv Respir Dis. 2016;10(5):468–484. doi:10.1177/1753465816654873.
- Zhang W, Nie Y, Chong L, Cai X, Zhang H, Lin B, Liang Y, Li C. PI3K and Notch signal pathways coordinately regulate the activation and proliferation of T lymphocytes in asthma. Life Sci. 2013;92(17-19):890–895. doi:10.1016/j.lfs.2013.03.005.
- Zhang W, Zhang X, Sheng A, Weng C, Zhu T, Zhao W, Li C. γ-secretase inhibitor alleviates acute airway inflammation of allergic asthma in mice by Downregulating Th17 cell differentiation. Mediators Inflamm. 2015;2015:258168–258167. doi:10.1155/2015/258168.
- Hu C, Li Z, Feng J, Tang Y, Qin L, Hu X, Zhang Y, He R. Glucocorticoids modulate Th1 and Th2 responses in asthmatic mouse models by inhibition of Notch1 signaling. Int Arch Allergy Immunol. 2018;175(1-2):44–52. doi:10.1159/000485890.
- Zhou M, Cui ZL, Guo XJ, Ren LP, Yang M, Fan ZW, Han RC, Xu WG. Blockade of notch signaling by γ-secretase inhibitor in lung T cells of asthmatic mice affects T Cell Differentiation and Pulmonary Inflammation. Inflammation. 2015;38(3):1281–1288. doi:10.1007/s10753-014-0098-5.
- Li C, Sheng A, Jia X, Zeng Z, Zhang X, Zhao W, Zhang W. Th17/Treg dysregulation in allergic asthmatic children is associated with elevated notch expression. J Asthma. 2018;55(1):1–7. doi:10.1080/02770903.2016.1266494.
- Rayees S, Malik F, Bukhari SI, Singh G. Linking GATA-3 and interleukin-13: implications in asthma. Inflamm Res. 2014;63(4):255–265. doi:10.1007/s00011-013-0700-6.
- Fang TC, Yashiro-Ohtani Y, Del Bianco C, Knoblock DM, Blacklow SC, Pear WS. Notch directly regulates Gata3 expression during T helper 2 cell differentiation. Immunity. 2007;27(1):100–110. doi:10.1016/j.immuni.2007.04.018.
- Caramori G, Lim S, Ito K, Tomita K, Oates T, Jazrawi E, Chung KF, Barnes PJ, Adcock IM. Expression of GATA family of transcription factors in T-cells, monocytes and bronchial biopsies. Eur Respir J. 2001;18(3):466–473. doi:10.1183/09031936.01.00040701.
- Nakamura Y, Ghaffar O, Olivenstein R, Taha RA, Soussi-Gounni A, Zhang DH, Ray A, Hamid Q. Gene expression of the GATA-3 transcription factor is increased in atopic asthma. J Allergy Clin Immunol. 1999;103(2 Pt 1):215–222. doi:10.1016/s0091-6749(99)70493-8.
- Zhang DH, Yang L, Cohn L, Parkyn L, Homer R, Ray P, Ray A. Inhibition of allergic inflammation in a murine model of asthma by expression of a dominant-negative mutant of GATA-3. Immunity. 1999;11(4):473–482. doi:10.1016/s1074-7613(00)80122-3.
- Liu, Ying, Zhou, Liting, Wu, Hanlin, Wang, Yitong, Zhang, Bo, Danzengluobu, . Role of notch signaling pathway in Muc5ac secretion induced by atmospheric PM2.5 in rats. Ecotoxicol Environ Saf 2022; 229: 113052. doi:10.1016/j.ecoenv.2021.113052.
- Liu Y, Zhou T, Sun L, Wang H, Zhou L. The effect of notch signal pathway on PM2.5-induced Muc5ac in Beas-2B cells. Ecotoxicol Environ Saf. 2020;203:110956. doi:10.1016/j.ecoenv.2020.110956.
- Wu JR, He Z, Bao HR, Zeng XL, Liu XJ. Study on the mechanism of PM2.5 affecting Th1/Th2 immune imbalance through the notch signaling pathway in asthmatic mice. Toxicol Res . 2023;12(4):675–684. doi:10.1093/toxres/tfad044.
- Herath K, Kim HJ, Mihindukulasooriya SP, Kim A, Kim HJ, Jeon YJ, Jee Y. Sargassum horneri extract containing mojabanchromanol attenuates the particulate matter exacerbated allergic asthma through reduction of Th2 and Th17 response in mice. Environ Pollut. 2020;265(Pt B):114094. doi:10.1016/j.envpol.2020.114094.
- Cheng M, Wang B, Yang M, Ma J, Ye Z, Xie L, Zhou M, Chen W. microRNAs expression in relation to particulate matter exposure: a systematic review. Environ Pollut. 2020;260:113961. doi:10.1016/j.envpol.2020.113961.
- Zhang HD, Jiang LH, Sun DW, Li J, Tang JH. MiR-139-5p: promising biomarker for cancer. Tumour Biol. 2015;36(3):1355–1365. doi:10.1007/s13277-015-3199-3.