Abstract
The emergency response following hurricanes Katrina and Rita included massive pumping and aeration operations to reduce biochemical oxygen demand (BOD) of floodwaters trapped within the city of New Orleans. Such engineering operations aerosolized tremendous quantities of water as microdroplets, which in turn increased the potential for waterborne microorganisms to partition into the atmosphere. To determine if remediation efforts significantly impacted local airborne microbe populations, or resulted in aerosolization of potentially pathogenic microorganisms, we performed direct microscopy, broad spectrum PCR, and DNA sequencing analysis on paired air and water samples collected in the immediate vicinity of turbulent pumping and aeration operations throughout flooded New Orleans. We report here that remediation activities following Hurricanes Katrina and Rita did not significantly impact bioaerosol ecology proximal to large engineering works, which promoted floodwater aerosolization. With exception to the minor representation of species associated with common skin infections, no pathogenic species were detected in this atmospheric sampling campaign. When compared to the growing genetic catalogues of atmospheric molecular ecology surveys, results from this sampling campaign were consistent with, but limited to, phylum level taxonomic patterns emerging from observations of outdoor bioaerosol communities.
1. INTRODUCTION
In the weeks following Hurricanes Katrina and Rita, the US Army Corps of Engineers drained nearly a trillion gallons of floodwater from the city of New Orleans by coordinating a massive pumping operation (USACE 2005a, b, c). Two water quality studies conducted immediately following Hurricane Katrina found elevated numbers of pathogenic bacteria (or cultured surrogate indicators) in floodwaters distributed throughout the city (CitationPardue et al. 2005; CitationPresley et al. 2006). High biochemical oxygen demands (BOD) (80 mg/L average) observed in many flooded locations, prompted emergency response crews to execute in-situ water reclamation practices using high-power mechanical aeration and turbulent pumping discharges in many of New Orleans' major canals. Such practices were employed for the express purpose of entraining air into moving floodwaters, and resulted in the aerosolization of tremendous quantities of water. Since significant numbers of microbes have been shown to partition into the atmosphere from relatively quiescent water bodies (CitationBlanchard and Syzdek 1970, Citation1971, Citation1972, Citation1982; CitationAngenent et al. 2005; CitationPaez-Rubio et al. 2005), the Katrina floodwater reclamation practices could have significantly impacted local aerobiology by enhancing the partitioning potential of waterborne microbes into aerosols.
Throughout New Orleans, floodwaters containing elevated numbers of potentially pathogenic microbes and high organic carbon levels were subject to sustained mechanical energy inputs on an enormous scale. In response to these unique environmental conditions, we identified and enumerated microorganisms associated with active water reclamation operations to determine whether the local ecology of airborne microbes was influenced by proximal water sources, and to ascertain if microbes present in the air posed a potential health risk to emergency response personnel working in the vicinity of water aerosolizing devices. Direct microscopy was used to quantify the microbiological loads present in aerosols and water samples. Broad-spectrum PCR and rRNA sequence analysis were conducted to assess microbial community compositions in the vicinity of outdoor remediation operations in the weeks between Hurricanes Katrina and Rita (September 18–September 25, 2005), and the week immediately following Rita's landfall.
2. MATERIALS AND METHODS
2.1. Aerosol and Source Water Sampling
A total of 55 samples were collected from 22 sites throughout the City of New Orleans between September 18 and 25, 2005. Based on safe accessibility, seven of these were chosen to collect concomitant aerosol and proximal source water samples in the immediate vicinity (not more than 2 m) of operating aerators and/or the turbulent terminus of floodwater transmission pipes. lists their GPS coordinates.
TABLE 1 Site identification key with GPS coordinates of the sampled locations in the flooded City of New Orleans during September, 2005. Three letter acronyms identify each site and timing of sample collection
As a cohort of unperturbed background observations, samples were also collected on the quiescent west shore of Lake Pontchartrain, where no engineering operations were engaged. During the entirety of this sampling campaign, local wind speeds were below measurable levels, which was the prevailing condition between Hurricanes Katrina and Rita.
Aerosol samples were collected with SKC Biosamplers (BioSampler SKC Inc., Eighty Four, PA), positioned approximately 1 meter above the ground. The collection medium was 20 ml of DNA-free sterile water (USP, Hospira, Inc. Lake Forest, IL). Biosamplers were operated at a flow rate of 12.5 L/min, at which they have a capture efficiency in excess of 90% for particles with mean aerodynamic diameters in the range between 0.4 and 10 μ M. Two impingers were run in parallel for each sampling site and collected continuously for at least one hour at each sampling site, in each case collecting a minimum air volume of 0.75 m3. To maintain appropriate aerosol collection efficiency within the impingers, sterile water was episodically added to the Biosamplers to make up for any evaporative losses. At the end of each sampling event, the collection fluids from each impinger were combined into DNA-free sterile 50 ml conical vials and placed on ice until they could be processed, which was less than 8 hours in all cases. A 1 liter grab sample of source water was collected at each of the sampling sites, approximately 30 min into each of the local aerosol sampling operations. Water samples were collected in sterile Nalgene bottles and stored on ice until they too could be similarly processed (< 8 h) as detailed in the following section.
Samples were processed and archived in the laboratory facilities at the Louisiana State University Coastal Ecology Institute. Samples were aliquoted as follows: 10 mL for direct microscopy analysis preserved with 2% (v/v) formaldehyde, and 30 mL (aerosol samples) or 990 mL (source water samples) for molecular analysis concentrated on 0.2 micrometer nitrocellulose filters (100 mL) (Nalgene Analytical Test Filter Funnel, Nalge Nunc International Rochester, NY). At the end of the sampling campaign all samples were stored on ice or frozen with liquid nitrogen and shipped overnight to the University of Colorado where they were stored at 4°C (direct count samples) or −20°C (DNA samples) until analyzed.
2.2. Bacterial Enumeration
Aerosol samples were stained and enumerated using 4′6-diamidino-2-phenylindole (DAPI) (Sigma Chemicals, St. Louis, MO) in accordance with previously described methods (CitationHernandez et al. 1999). DAPI is a nonspecific fluorescent DNA intercalating agent that has been successfully used for quantitative microscopy in other laboratory and environmental aerosol applications (CitationPaez-Rubio et al. 2005); it was used as described here with relative low background fluorescence observed in air or water samples. A minimum of 10 random fields were counted per slide and only intact, brightly stained cells with obvious bacterial or fungal morphology were counted. Direct counts were reported as the average of all fields counted; in all cases the coefficient of variation observed was less than 20%.
2.3. Genomic DNA Extraction
Concentrated samples and negative controls were extracted from the filters using a bead beating protocol modified from CitationFrank et al. (2003). Modifications to the protocol were made due to the material nature of the nitrocellulose filter. Briefly, filtered samples were dissolved using a bead beating solution containing ethyl acetate prior to the chloroform extraction process. Extracted DNA was resuspended using 20 microliters of Tris-Ethylenediamine Tetraacetic Acid (TE). Extracted DNA concentrations were measured using a PicoGreen dsDNA Quantitation Kit (Molecular Probes, Carlsbad, CA) and the NanoDrop® ND-3300 Fluorospectrometer (NanoDrop Technologies, Wilmington, DE).
2.4. PCR of rRNA Genes
Extracted DNA was assayed using universally conserved (515F/1391Rev) and bacteria specific (8F/1391Rev) primers following the protocol described by Papineau and coworkers (CitationPapineau et al. 2005). PCR controls with no DNA template were included with every assay. Extraction and negative controls never showed amplification.
2.5. Clone Library Construction
Clone libraries were constructed for each of the primer pairs (bacterial and universal) used. Each library was constructed of at least 96 randomly selected rRNA clones using a widely accepted method previously described by Papineau and coworkers (2005).
2.6. DNA Sequencing
Sequencing was performed as previously described by CitationPapineau et al. (2005). Base calling and assembly of raw sequence data were performed with the PHRED and PHRAP software packages (CitationEwing et al. 1998) using the software XplorSeq (CitationFrank 2008).
2.7. Phylogenetic Analysis
The sequence collection was screened for chimeric sequences with the Bellerophon software package (CitationHuber et al. 2004). The closest known relative of each rRNA clone was determined by comparison of its sequence to all rRNA sequences in GenBank by BLAST (CitationAltschul et al. 1990). Sequences were aligned with the NAST software package (CitationDeSantis et al. 2006) and their phylogenetic positions were determined by analysis with the ARB software package (CitationLudwig et al. 2004) and a curated ARB database and tree with ∼ 100,000 rRNA sequences ≥ 1250 nucleotides in length from the NAST project (CitationDeSantis et al. 2006). Sequences were added to the ARB tree by parsimony insertion.
2.8. Richness Estimates
Estimates of potential true rRNA sequence diversity in each sample were calculated with the Chao1 statistic (CitationChao 1984) as previously described (CitationPapineau et al. 2005). Sequences were collected into operational taxonomic units (OTUs), or relatedness groups, based on sequence identity at fixed intervals ranging from 95% to 100% in increments of 1%. OUT data were used to calculate the Chao1 richness estimate using a Python script (J.J. Walker, unpublished software).
2.9. Comparative Phylogenetic Methods
Sequence sets representative of the microbial composition of water and air samples were compared statistically with the online phylogenetic statistics software package UniFrac (CitationLozupone and Knight 2005; CitationLozupone et al. 2006) and the SONS program (CitationSchloss and Handelsman 2006). UniFrac compares sets of related sequences (samples) using a phylogenetic tree and calculates a comparative metric based on the fraction of the total distance in the tree that is unique to a particular sample. Relationships between samples were explored using the distance metric with two complementary methods—hierarchical classification and ordination. Samples were clustered hierarchically with the Unweighted Pair Group Method with Arithmetic means (UPGMA), and statistical support for the resulting phylogenetic tree was assessed by jackknife iterations. Principal Coordinates Analysis (PCA) (CitationLozupone et al. 2007) was used to order the samples by maximizing the linear correlation between distances from the UniFrac distance matrix in a two dimensional projection. The SONS program compared the membership and structure of the selected (sourced) communities at a specified phylogenetic level. Based on the selected phylogenetic level, the program assigned sequences to operation taxonomic unit (OTU) bins (membership) and then evaluated the abundance in each bin for each sample and associated community content. Community similarity was assessed based on shared and unique memberships and overall extent of structure alikeness.
2.10. Potential Pathogens
A list of known human pathogens was used to compare the closest match by BLAST for all 2100 sequences catalogued in this study (CitationTaylor et al. 2001). The list of pathogen names and the closest BLAST matches were exported to Microsoft Excel and compared using a series of lookup functions.
3. RESULTS
3.1. Microbe Quantitation
As determined by direct epifluorescent microscopy, the concentrations of bacteria, fungi and their spores in the floodwater and aerosol samples are summarized in .
FIG. 1 Direct microscopic count of bacteria, fungi, and their spores in atmospheric samples and nearby aerated/turbulent floodwaters. Left scale refers to source water concentrations; right scale refers to aerosol concentrations.
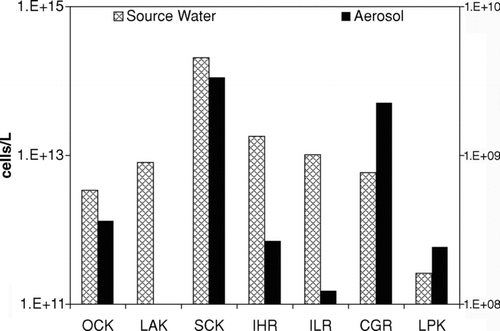
Floodwater microbe concentrations were ten-fold or more over that observed in western Lake Pontchartrain, the designated background, and were similar to microbial numbers previously reported for partially treated domestic wastewater (2 × 1011 cells/L) (CitationPaez-Rubio et al. 2005). The bioaerosol loads measured above the Katrina flood zone were in the range between 1 × 105 and 3 × 106 cell/m3; these levels were elevated above those reported for other undisturbed outdoor environments (2 × 104 − 1 × 105 cell/m3) (CitationTong and Lighthart 1999; CitationMaron et al. 2005) and were similar to those measured in another flood-impacted region (La Junta, CO, 3 × 105 − 2 × 107 cell/m3) (CitationFabian et al. 2005); these levels however, were markedly less than those reported from a Sonoran desert site flooded with partially treated domestic wastewater (Chihuahua, Mexico, 1 × 107 − 1 × 109 cell/m3) (CitationPaez-Rubio et al. 2005). Based on the data available from the aforementioned bioaerosol studies using similar sampling equipment and microscopic analyses, the airborne microbe concentrations observed here are in the range of other outdoor bioaerosol concentrations observed in the absence of measurable wind velocity.
3.2. Microbial Composition and Relative Representation
DNA extracted from the collected samples was used as templates for PCR with both bacterial (8F/1391R) and universal (515F/1391R) primers. Resultant PCR products then were processed such that a minimum of 96 randomly selected clones were sequenced for each sample. A total of 28 rRNA gene libraries (both bacterial and universal) were analyzed and 2100 sequences determined. summarizes the characteristics associated with these clone libraries.
TABLE 2 Summary of clone library characteristics for each sample (B corresponds to those libraries generated with Bacterial primer sets; U corresponds to those libraries generated with Universal primer sets)
When possible composite (bacterial and universal) libraries were used for phylogenetic analyses. While this sampling and analysis scheme did not have the statistical power to detect all phylotypes, it provided a robust survey of the more dominant taxonomic units. summarizes the phylogenetic distribution of rRNA sequences observed in the proximal air and water environments, to which nonparametric Chao1 analyses was applied to estimate richness (CitationChao 1984) within the collected samples.
FIG. 2 Phylogenetic distribution of rRNA sequences observed from pooled atmospheric samples and nearby aerated/turbulent floodwaters.
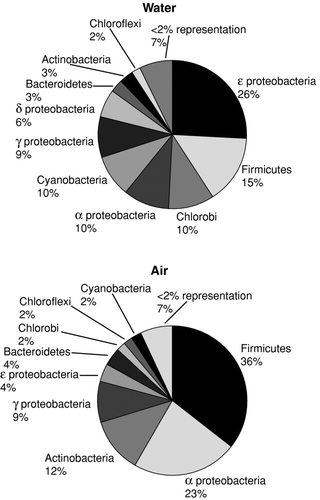
Sequences were parsed into relatedness groups based on operational taxonomic units (OTUs) assigned to differentiate species at or above 97% sequence identity. As judged by this level of stringency, between 55 and 390 OTUs were estimated to populate the air samples, where proximal water samples were predicted to contain between 190 and 470 distinct OTUs. A summary of the Chao results are listed in . Chao1 analyses as defined by this OTU level suggested that all but one of the 14 samples analyzed contained a range of bacterial diversity that was beyond the resolution of the clone libraries observed here. However, more than 90% of the sequences resolved were associated with 10 of the currently indexed bacterial phyla, and no sequences recovered were associated known human pathogens.
3.3. Atmospheric–Aquatic Relationships
To assess potential relationships between local airborne and aquatic community structure, we compared their phylogenetic composition using accepted bioinformatics practices leveraging UniFrac analyses with an Unweighted Pair Group Method with Arithmetic Mean (UPGMA) (CitationLozupone and Knight 2005). These were applied to quantify relationships between different OTU sets in a phylogenetic tree built solely from these greater New Orleans samples; an independent principal coordinates analysis (PCA) was then applied to expose salient environmental associations.
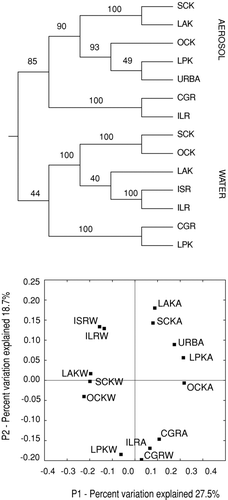
Results of the UPGMA analysis summarized in , showed a distinct separation between aerosol and proximal source water sequences, which was statistically supported by Jackknife permutations (n > 1000). In all cases, OTU clusters in aerosol samples were more similar to one another than to OTU clusters in proximal floodwaters. PCA converged on a complimentary result (), which suggested that the phylogenetic variations observed could be explained by source—atmosphere or floodwater—and that some variation could be associated with collection date (pre– or post–Hurricane Rita). Inclusion of sequence data (URBA) from an unrelated atmospheric aerosol study (CitationBrodie et al. 2007) in the UPGMA and PCA analyses did not alter aerosol and source water clustering ().
FIG. 3 Acronyms identify each site and timing of sample collection. Sample designations including K, signify sampling date between hurricanes Katrina and Rita; sample designation including R signifies sampling date within a week of Rita's landfall. Final letter signifies sampling environment: designations ending with A, signify atmospheric sample; designations ending with W signify floodwater sample. The sample identified as URBA represents sequence data from an unrelated aerosol study by CitationBrodie et al. (2007). (Top) UPGMA distance matrix results for sequenced samples recovered from air and proximal floodwaters environments following 1000 jackknife iterations. (Bottom) Principal Components Analysis (PCA); Component 1 (sample environment) is on the x-axis. Component 2 (sample date) is on the y-axis.
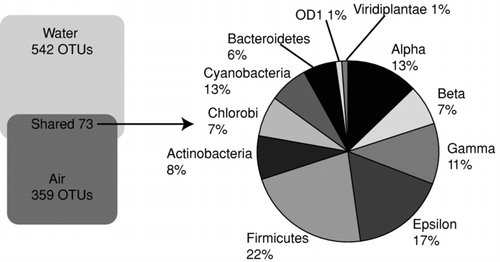
UniFrac analyses did not identify any significant correlations between the conglomerate phylogenetic compositions of the paired air and water samples; however, some phyla overlap suggested some sites may have had similar kinds of organisms. In order to assess this, we inspected the OTUs shared among samples using the SONS analysis package (CitationSchloss and Handelsman 2006), which binned these sequence collections into OTUs. and summarize SONS outcomes, showing that only 73 of the total 974 OTUs observed, were shared between the atmospheric and floodwater samples. Further, four of seven sites had less than 8% of their OTUs present any significant overlap in proximal air and water samples; the maximum overlap at any given site was approximately 20% of the total OTUs recovered. provides a listing of the OTUs shared between the proximal water and air samples.
TABLE 3 Summary of shared species between air and proximal water samples, and the frequency of their occurrence. Sites CGR and OCK are not included because no OTUs were shared between their air and water samples
FIG. 4 Percent distribution of OTUs (>97% sequence identity) as judged by rRNA sequences observed in atmospheric samples and nearby aerated/turbulent floodwaters. A total of 954 distinct OTUs were observed in both the air and water environments. While 73 OTUs were shared between the two environments some sites had almost no overlap; other shared up to 18% overlap.
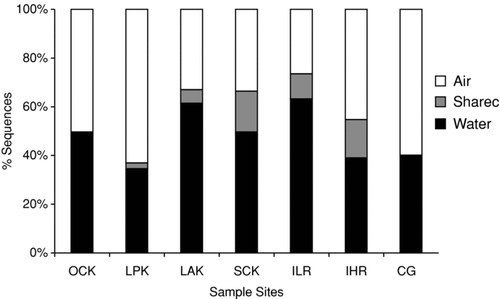
FIG. 5 Sequence associations of the 73 shared OTUs (>97% sequence identity) in atmospheric samples and nearby aerated/turbulent floodwaters.
Closer examination of the shared OTUs revealed that in most instances the shared sequences were not from the same sampling environment. Community relatedness among the air samples was also examined using SONS analysis. Samples collected prior to Hurricane Rita (designated with K) shared 2 OTUs—Mangroveibacter plantisponsor and Exiguobacterium sp.—that combined to account for less than 2% of the total sequence data recovered from the environments considered. A similar finding was observed for the three samples collected following Hurricane Rita (designated with R). Variovorax paradoxus was the only species common to all three environments and accounted for a total of 5 sequences (< 1% of the total sequence data for the three environments considered). No OTU was common to all the aerosol environments.
Regional Aerosol Communities
The relatedness between air samples and lack of significant overlap between OTUs recovered from aerosols and nearby aerated/turbulent floodwaters prompted indications of an intraregional aerobiological population with respect to the near-ground level atmosphere. The concept of an ubiquitous aerosol community was explored through additional SONs and Unifrac analyses using additional atmospheric sequence data available in GenBank (CitationHughes et al. 2004; CitationMaron et al. 2005; CitationPaez-Rubio et al. 2005; CitationBaertsch et al. 2007; CitationBrodie et al. 2007; CitationFierer et al. 2008). When scrutinized at species level (97% sequence ID), SONS analysis indicated few examples of shared diversity. A total of 1530 OTUs were present among the different aerosol environments. Of these OTUs, 229 were shared between any two environments and only 77 included sequences from the Katrina data set. These 77 OTUs accounted for 16% of the total Katrina aerosol sequence data, with a quarter of these sequences being observed only once. At the less stringent level (phylum), many of the atmospheric environments ostensibly had similarities in their composition, with representatives of the Proteobacteria, Firmicutes, and Actinobacteria dominating these sequence databases. UniFrac analyses applied to seek order among these atmospheric databases were inconclusive because of variability. Such variance between environments could be the result of many different factors including the numbers of sequences considered, metrological factors, and sample collection differences.
DISCUSSION
Relatively little is known about the kinds of organisms that populate the atmosphere on local, regional or continental scales (CitationPeccia and Hernandez 2006). Aerobiology studies have traditionally focused on health concerns associated with indoor environments, with particular emphasis on airborne fungi and their spores (CitationHambraeus 1988; CitationBurge 1990; CitationMadelin 1994; CitationHorner et al. 1995; CitationGorny et al. 2002), and there is a paucity of outdoor bioaerosol surveys. While not a large survey, this study adds to the growing inventory of seasonal observations of atmospheric microbial communities, and does so through a unique environmental window: a flooded and deserted metropolitan Gulf Coast city, during the windless September weeks between Hurricanes Katrina and Rita.
Much of our knowledge about the microbes that inhabit the atmosphere has been acquired from culture-based studies, even though it is established that culture methods are biased and insufficient for capturing the microbial diversity present in most environments (CitationAmann et al. 1995; CitationPace 1997). Moreover, characterization of airborne populations is difficult due to relatively low cell densities and local meteorological challenges. In recent years, reliable culture independent methods have been developed (CitationAlvarez et al. 1995; CitationPeccia and Hernandez 2006) and utilized for the characterization and quantification of bioaerosols.
While modern molecular methods, in particular PCR-based surveys, alleviate biases associated with microbial recovery by culture, and greatly increase sampling sensitivity, they have their own limitations. These include the biases associated with target amplicons, primer annealing, polymerase error, annealing temperatures, cycle number and template concentration, and the number of primer pairs used (CitationFarrelly et al. 1995; CitationSuzuki and Giovannoni 1996; CitationPolz and Cavanaugh 1998; CitationBecker et al. 2000; CitationIshii and Fukui 2001; CitationHongoh et al. 2003; CitationAcinas et al. 2005; CitationOsborne et al. 2005; CitationSipos et al. 2007; CitationJeon et al. 2008; CitationHuber et al. 2009). These shortcomings skew our understanding of the diversity present within any environmental sample and the atmosphere is certainly no exception.
In addition to methodologically induced limitations, other limitations of this work include its intrinsic nature of its irreproducibility (post–natural disaster New Orleans) and limited sample and sequence data sizes. The cleanup efforts involved in restoring the city were quick and comprehensive. Flooded neighborhoods were systematically drained in the course of a day making repeat sampling impossible. Likewise, the loss of infrastructure, such as bridges and roads, as well as the massive amount of debris, such as boats, cars, and trees, made navigating the city to arrive at sampling locations difficult at best. In many instances sites of intense remediation efforts were only accessible by military helicopter.
The total bioaerosol loads observed in this study were similar to those reported for another flood impacted landscape (CitationFabian et al. 2005), but were considerably less (100–200×) than those published for site flooded with domestic wastewater (CitationPaez-Rubio et al. 2005). The microbial loads measured in the proximal source water samples were similar to those previously reported for domestic wastewater. With the extensive energy input from remediation activities and relatively elevated source water microbial concentrations, the local bioaerosol loads may have been expected to be greater. The disparity in microbial concentrations between the source water and local aerosol samples may suggest limited partitioning of waterborne microbes to the local air. Weather conditions (high humidity and negligible wind), which prevailed during this sampling campaign, may have impacted these physical partition rates (CitationHarrison et al. 2005; CitationMaron et al. 2006). Bioaerosol loads measured near the remediation activities appear to be characteristic of what is emerging to be a relatively consistent range of outdoor near-ground levels given the current catalogue of bioaerosol studies executing direct microbial counting (CitationTong and Lighthart 1999; CitationAngenent et al. 2005; CitationMaron et al. 2005; Rodriguez de Evgrafov, unpublished data).
As judged by modern phylogenetics, the microbial composition of floodwaters and the associated local atmospheric environments presented relatively limited diversity, where 10 phyla circumscribed more than 90% of the total sequences recovered.
Both the atmospheric environment and some floodwaters appeared to be significantly influenced by the passage of Hurricane Rita. The aerosol contents were markedly different for samples collected between hurricanes Katrina and Rita than those collected immediately after Rita. Prior to Hurricane Rita, representatives of the Firmicutes comprised 41% of the overall airborne ecology, mainly spore-forming Bacillus spp. and Clostrium spp., with α -proteobacteria representatives being the next largest group. Following Hurricane Rita, however, representatives of Firmicutes disappeared almost entirely and the α -proteobacteria representatives constituted 33% of the total atmospheric sequences—under otherwise identical meteorological conditions. A similar division was also observed in the water samples. ε -proteobacteria representatives composed the largest group of organisms in the sampled water bodies prior to Hurricane Rita; however, after the storm event, the dominant phyla in the flood waters shifted to representatives of the Firmicutes. Of the ε -proteobacteria representatives observed, nearly half were members of the Sulfurimonaceace family, perhaps indicating that anoxic conditions dominated the sampled water bodies at that time. Firmicutes observed in water samples following Hurricane Rita were primarily Erysipelothrix spp., related to organisms cultured from a mangrove and an anoxic swine lagoon.
Recent studies suggest that the risk for airborne infectious disease outbreak is low following natural disasters, particularly for developed nations (CitationShultz et al. 2005; CitationFloret et al. 2006). Most acute medical cases reported during the first three weeks following Katrina were dermatologic in nature (CDC 2005). While incidences of upper respiratory infections and pneumonias were also reported, there were no major reported respiratory disease outbreaks (CDC 2005; CitationSinger 2005), and a medical group reported that management of chronic illnesses made up most of their patient visits (CitationCurrier et al. 2006). We report here that sequences corresponding to Propionibacterium acnes and Staphylococcus epidermidis were the two most abundant potentially pathogenic organisms with greater than 97% BLAST ID. These results are consistent with the medical reports from the Katrina disaster, and do not support the hypothesis that there was an elevated health risk to emergency response personnel in this environment from flood remediation activities.
Richness estimates applied to each sample determined that almost all environments were under sampled given the resolution offered by the clone libraries recovered. These richness findings were not unexpected given the dynamic environmental events and cleanup operations that took place during the sampling campaign, and the numbers of sequences recovered and analyzed. Even so, the microbial compositions observed provide some understanding of the types and distribution of microorganisms that were present in these unique environments.
Results from community comparisons with UniFrac (), which incorporated sequence data from another large aerosol study (CitationBrodie et al. 2007), showed that microbes observed in the New Orleans near ground atmosphere were more similar to an extra-regional aerosol environment than they were to their proximal source waters. Likewise a SONS community membership analysis of the post-Katrina air and water sequences, which revealed that less than 10% of species-level OTUs were shared between the two environments, and those in common were not from the same environment in most instances. Similar analyses among the air samples revealed few similarities among the sampled environments. These results may suggest that the source of the recovered aerosol sequences was the local atmospheric community rather than the corresponding source waters.
Disconnect between the dominant microbes found in the local atmosphere and perturbed floodwaters in this setting may indicate that the airborne microbes detected are part of a complex collection of organisms that may pervade near ground-level on a yet-to-be determined environmental scale. Indeed, microbial physiology and water quality factors can influence the potential for microbes (and chemicals) to transfer from water sources into aerosols (CitationAngenent et al. 2005; CitationPaez-Rubio et al. 2005; CitationBaertsch et al. 2007). In the samples analyzed however, any contribution to atmospheric biocomplexity from artificially generated aerosols appears minor, regardless of the tremendous energy inputs from mechanical aerators and pump-induced turbulence. These results suggest that the combination to microbial physiology and environmental conditions did not promote the aerosol partitioning of waterborne microbes.
Through a series of additional statistical analyses leveraging sequence data from other airborne molecular ecology surveys, we observed that membership similarity among these atmospheric environments was limited to a phylum level. Community structure analyses at more stringent phylogenetic levels confirmed that the membership similarity in genetically characterized atmospheric communities observed is superficial and there likely exists great variability among the airborne microbial ecology in different atmospheric environments; the spatial and temporal scale is yet to be determined. This conclusion is not surprising considering the dynamic nature of the atmosphere, sampling and analytical differences between this and other atmospheric molecular studies, and the limited genetic data available for analyses. Additional long term molecular based aerosol studies are needed to resolve the environmental scales germane to the airborne microbial biosphere.
REFERENCES
- Acinas , S. G. , Sarma-Rupavtarm , R. , Klepac-Ceraj , V. and Polz , M. F. 2005 . PCR-Induced Sequence Artifacts and Bias: Insights from Comparison of Two 16S rRNA Clone Libraries Constructed from the Same Sample . Appl. Environ. Microbiol. , 71 : 8966 – 8969 .
- Altschul , S. F. , Gish , W. , Miller , W. , Myers , E. W. and Lipman , D. J. 1990 . Basic Local Alignment Search Tool . J. Mol. Biol. , 215 : 403 – 410 .
- Alvarez , A. J. , Buttner , M. P. and Stetzenbach , L. D. 1995 . PCR for Bioaerosol Monitoring—Sensitivity and Environmental Interference . Appl. Environ. Microbiol. , 61 : 3639 – 3644 .
- Amann , R. I. , Ludwig , W. and Schleifer , K. H. 1995 . Phylogenetic Identification and In-Situ Detection of Individual Microbial-Cells without Cultivation . Microbiol. Rev. , 59 : 143 – 169 .
- Angenent , L. T. , Kelley , S. T. , St Amand , A. , Pace , N. R. and Hernandez , M. T. 2005 . Molecular Identification of Potential Pathogens in Water and Air of a Hospital Therapy Pool . Proc. Natl. Acad. Sci. USA , 102 : 4860 – 4865 .
- Baertsch , C. , Paez-Rubio , T. , Viau , E. and Peccia , J. 2007 . Source Tracking Aerosols Released from Land-Applied Class B Biosolids During High-Wind Events . Appl. Environ. Microbiol. , 73 : 4522 – 4531 .
- Becker , S. , Boger , P. , Oehlmann , R. and Ernst , A. 2000 . PCR Bias in Ecological Analysis: A Case Study for Quantitative Taq Nuclease Assays in Analyses of Microbial Communities . Appl. Environ. Microbiol. , 66 : 4945 – 4953 .
- Blanchard , D. and Syzdek , L. 1970 . Mechanism for the Water-to-Air Transfer and Concentration of Bacteria . Science , 170 : 626 – 628 .
- Blanchard , D. and Syzdek , L. 1971 . Bubbles and Water-To Air Transfer of Bacteria . Bull. Amer. Meteorol. Soc. , 52 : 1136
- Blanchard , D. C. and Syzdek , L. D. 1972 . Concentration of Bacteria in Jet Drops from Bursting Bubbles . J. Geophys. Res. , 77 : 5087 – 5099 .
- Blanchard , D. C. and Syzdek , L. D. 1982 . Water-to-Air Transfer and Enrichment of Bacteria in Drops from Bursting Bubbles . Appl. Environ. Microbiol. , 43 : 1001 – 1005 .
- Brodie , E. L. , DeSantis , T. Z. , Parker , J. P. M. , Zubietta , I. X. , Piceno , Y. M. and Andersen , G. L. 2007 . Urban Aerosols Harbor Diverse and Dynamic Bacterial Populations . Proc. Natl. Acad. Sci. USA , 104 : 299 – 304 .
- Burge , H. 1990 . Bioaerosols—Prevalence and Health-Effects in the Indoor Environment . J. Allergy and Clinical Immunology , 86 : 687 – 701 .
- CDC . 2005 . Infectious Disease and Dermatologic Conditions in Evacuees and Rescue Workers After Hurricane Katrina—Multiple States, August-September, 2005 . JAMA , 294 : 2158 – 2160 .
- Chao , A. 1984 . Nonparametric-Estimation of the Number of Classes in a Population . Scan. J. Statistics , 11 : 265 – 270 .
- Currier , M. , King , D. S. , Wofford , M. R. , Daniel , B. J. and deShazo , R. 2006 . A Katrina Experience: Lessons Learned . Amer. J. Medicine , 119 : 986 – 992 .
- DeSantis , T. Z. , Hugenholtz , P. , Keller , K. , Brodie , E. L. , Larsen , N. Piceno , Y. M. 2006 . NAST: A Multiple Sequence Alignment Server for Comparative Analysis of 16S rRNA Genes . Nucleic Acids Res. , 34 : W394 – W399 .
- Ewing , B. , Hillier , L. , Wendl , M. C. and Green , P. 1998 . Base-Calling of Automated Sequencer Traces Using phred. I. Accuracy Assessment . Genome Research , 8 : 175 – 185 .
- Fabian , M. P. , Miller , S. L. , Reponen , T. and Hernandez , M. T. 2005 . Ambient Bioaerosol Indices for Indoor Air Quality Assessments of Flood Reclamation . J. Aerosol Sci. , 36 : 763 – 783 .
- Farrelly , V. , Rainey , F. A. and Stackebrandt , E. 1995 . Effect of Genome Size and RRN Gene Copy Number on PCR Amplification of 16S Ribosomal-RNA Genes from a Mixture of Bacterial Species . Appl. Environ. Microbiol. , 61 : 2798 – 2801 .
- Fierer , N. , Liu , Z. Z. , Rodriguez-Hernandez , M. , Knight , R. , Henn , M. and Hernandez , M. T. 2008 . Short-Term Temporal Variability in Airborne Bacterial and Fungal Populations . Appl. Environ. Microbiol. , 74 : 200 – 207 .
- Floret , N. , Viel , J. F. , Mauny , F. , Hoen , B. and Piarroux , R. 2006 . Negligible Risk for Epidemics After Geophysical Disasters . Emerging Infectious Diseases , 12 : 543 – 548 .
- Frank , D. N. 2008 . XplorSeq: A Software Environment for Integrated Management and Phylogenetic Analysis of Metagenomic Sequence Data . BMC Bioinformatics , 9 : 16
- Frank , D. N. , Spiegelman , G. B. , Davis , W. , Wagner , E. , Lyons , E. and Pace , N. R. 2003 . Culture-Independent Molecular Analysis of Microbial Constituents of the Healthy Human Outer Ear . J. Clin. Microbiol. , 41 : 295 – 303 .
- Gorny , R. L. , Reponen , T. , Willeke , K. , Schmechel , D. , Robine , E. , Boissier , M. and Grinshpun , S. A. 2002 . Fungal Fragments as Indoor Air Biocontaminants . Appl. Environ. Microbiol. , 68 : 3522 – 3531 .
- Hambraeus , A. 1988 . Aerobiology in the Operating-Room—A Review . J. Hospital Infection , 11 : 68 – 76 .
- Harrison , R. M. , Jones , A. M. , Biggins , P. D. E. , Pomeroy , N. , Cox , C. S. , Kidd , S. P. , Hobman , J. L. , Brown , N. L. and Beswick , A. 2005 . Climate Factors Influencing Bacterial Count in Background Air Samples . Intl. J. Biometeorology , 49 : 167 – 178 .
- Hernandez , M. , Miller , S. L. , Landfear , D. W. and Macher , J. M. 1999 . A Combined Fluorochrome Method for Quantitation of Metabolically Active and Inactive Airborne Bacteria . Aerosol Sci. Technol. , 30 : 145 – 160 .
- Hongoh , Y. , Yuzawa , H. , Ohkuma , M. and Kudo , T. 2003 . Evaluation of Primers and PCR Conditions for the Analysis of 16S rRNA Genes from a Natural Environment . FEMS Microbiol. Lett. , 221 : 299 – 304 .
- Horner , W. E. , Helbling , A. , Salvaggio , J. E. and Lehrer , S. B. 1995 . Fungal Allergens . Clin. Microbiol. Rev. , 8 : 161 – 179 .
- Huber , J. A. , Morrison , H. G. , Huse , S. M. , Neal , P. R. , Sogin , M. L. and Welch , D. B. M. 2009 . Effect of PCR Amplicon Size on Assessments of Clone Library Microbial Diversity and Community Structure . Environ. Microbiol. , 11 : 1292 – 1302 .
- Huber , T. , Faulkner , G. and Hugenholtz , P. 2004 . Bellerophon: A Program to Detect Chimeric Sequences in Multiple Sequence Alignments . Bioinformatics , 20 : 2317 – 2319 .
- Hughes , K. A. , McCartney , H. A. , Lachlan-Cope , T. A. and Pearce , D. A. 2004 . A Preliminary Study of Airborne Microbial Biodiversity Over Peninsular Antarctica . Cellular and Molecular Biology , 50 : 537 – 542 .
- Ishii , K. and Fukui , M. 2001 . Optimization of Annealing Temperature to Reduce Bias Caused by a Primer Mismatch in Multitemplate PCR . Appl. Environ. Microbiol. , 67 : 3753 – 3755 .
- Jeon , S. , Bunge , J. , Leslin , C. , Stoeck , T. , Hong , S. H. and Epstein , S. S. 2008 . Environmental rRNA Inventories Miss Over Half of Protistan Diversity . BMC Microbiol. , 8 : 13
- Lozupone , C. , Hamady , M. , Kelley , S. T. and Knight , R. 2007 . Quantitative and Qualitative Beta Diversity Measures Lead to Different Insights into Factors that Structure Microbial Communities . Appl. Environ. Microbiol. , 73 : 1576 – 1585 .
- Lozupone , C. , Hamady , M. and Knight , R. 2006 . UniFrac—An Online Tool for Comparing Microbial Community Diversity in a Phylogenetic Context . BMC Bioinformatics , 7 : 371 – 385 .
- Lozupone , C. and Knight , R. 2005 . UniFrac: A New Phylogenetic Method for Comparing Microbial Communities . Appl. Environ. Microbiol. , 71 : 8228 – 8235 .
- Ludwig , W. , Strunk , O. , Westram , R. , Richter , L. , Meier , H. , Yadhukumar , Buchner , A. , Lai , T. , Steppi , S. , Jobb , G. , Forster , W. , Brettske , I. , Gerber , S. , Ginhart , A. W. , Gross , O. , Grumann , S. , Hermann , S. , Jost , R. , Konig , A. , Liss , T. , Lussmann , R. , May , M. , Nonhoff , B. , Reichel , B. , Strehlow , R. , Stamatakis , A. , Stuckmann , N. , Vilbig , A. , Lenke , M. , Ludwig , T. , Bode , A. and Schleifer , K. H. 2004 . ARB: A Software Environment for Sequence Data . Nucleic Acids Res. , 32 : 1363 – 1371 .
- Madelin , T. M. 1994 . Fungal Aerosols—A Review . J. Aerosol Sci. , 25 : 1405 – 1412 .
- Maron , P. A. , Lejon , D. P. H. , Carvalho , E. , Bizet , K. , Lemanceau , P. , Ranjard , L. and Mougel , C. 2005 . Assessing Genetic Structure and Diversity of Airborne Bacterial Communities by DNA Fingerprinting and 16S rDNA Clone Library . Atmos. Environ. , 39 : 3687 – 3695 .
- Maron , P. A. , Mougel , C. , Lejon , D. P. H. , Carvalho , E. , Bizet , K. , Marck , G. , Cubito , N. , Lemanceau , P. and Ranjard , L. 2006 . Temporal Variability of Airborne Bacterial Community Structure in an Urban Area . Atmos. Environ. , 40 : 8074 – 8080 .
- Osborne , C. A. , Galic , M. , Sangwan , P. and Janssen , P. H. 2005 . PCR-Generated Artefact from 16S rRNA Gene-Specific Primers . FEMS Microbiol. Lett. , 248 : 183 – 187 .
- Pace , N. R. 1997 . A Molecular View of Microbial Diversity and the Biosphere . Science , 276 : 734 – 740 .
- Paez-Rubio , T. , Viau , E. , Romero-Hernandez , S. and Peccia , J. 2005 . Source Bioaerosol Concentration and rRNA Gene-Based Identification of Microorganisms Aerosolized at a Flood Irrigation Wastewater Reuse Site . Appl. Environ. Microbiol. , 71 : 804 – 810 .
- Papineau , D. , Walker , J. J. , Mojzsis , S. J. and Pace , N. R. 2005 . Composition and Structure of Microbial Communities from Stromatolites of Hamelin Pool in Shark Bay, Western Australia . Appl. Environ. Microbiol. , 71 : 4822 – 4832 .
- Pardue , J. H. , Moe , W. M. , McInnis , D. , Thibodeaux , L. J. , Valsaraj , K. T. , Maciasz , E. , van Heerden , I. , Korevec , N. and Yuan , Q. Z. 2005 . Chemical and Microbiological Parameters in New Orleans Floodwater Following Hurricane Katrina . Environ. Sci. Technol. , 39 : 8591 – 8599 .
- Peccia , J. and Hernandez , M. 2006 . Incorporating Polymerase Chain Reaction-Based Identification, Population Characterization, and Quantification of Microorganisms into Aerosol Science: A Review . Atmos. Environ. , 40 : 3941 – 3961 .
- Polz , M. F. and Cavanaugh , C. M. 1998 . Bias in Template-to-Product Ratios in Multitemplate PCR . Appl. Environ. Microbiol. , 64 : 3724 – 3730 .
- Presley , S. M. , Rainwater , T. R. , Austin , G. P. , Platt , S. G. , Zak , J. C. , Cobb , G. P. , Marsland , E. J. , Tian , K. , Zhang , B. H. , Anderson , T. A. , Cox , S. B. , Abel , M. T. , Leftwich , B. D. , Huddleston , J. R. , Jeter , R. M. and Kendall , R. J. 2006 . Assessment of Pathogens and Toxicants in New Orleans, LA Following Hurricane Katrina . Environ. Sci. Technol. , 40 : 468 – 474 .
- Schloss , P. D. and Handelsman , J. 2006 . Introducing SONS, a Tool for Operational Taxonomic Unit-Based Comparisons of Microbial Community Memberships and Structures . Appl. Environ. Microbiol. , 72 : 6773 – 6779 .
- Shultz , J. M. , Russell , J. and Espinel , Z. 2005 . Epidemiology of Tropical Cyclones: The Dynamics of Disaster, Disease, and Development . Epidemiologic Reviews , 27 : 21 – 35 .
- Singer , E. 2005 . Katrina Disease Fears Unfounded . Nature Medicine , 11 : 1015 – 1015 .
- Sipos , R. , Szekely , A. J. , Palatinszky , M. , Revesz , S. , Marialigeti , K. and Nikolausz , M. 2007 . Effect of Primer Mismatch, Annealing Temperature and PCR Cycle Number on 16S rRNA Gene-Targetting Bacterial Community Analysis . FEMS Microbiol. Ecol. , 60 : 341 – 350 .
- Suzuki , M. T. and Giovannoni , S. J. 1996 . Bias Caused by Template Annealing in the Amplification of Mixtures of 16S rRNA Genes by PCR . Appl. Environ. Microbiol. , 62 : 625 – 630 .
- Taylor , L. H. , Latham , S. M. and Woolhouse , M. E. J. 2001 . Risk Factors for Human Disease Emergence . Philosophical Transactions of the Royal Society of London Series B-Biological Sciences , 356 : 983 – 989 .
- Tong , Y. Y. and Lighthart , B. 1999 . Diurnal Distribution of Total and Culturable Atmospheric Bacteria at a Rural Site . Aerosol Sci. Technol. , 30 : 246 – 254 .
- USACE . 2005a . Corps Marks Halfway Point in Unwatering Mission , U.S. Army Corps of Engineers, New Orleans District Public Affairs .
- USACE . 2005b . Corps of Engineers' Update to Hurricane Katrina Response , U.S. Army Corps of Engineers, Mississippi Valley Division Public Affairs .
- USACE . 2005c . New Orleans Water Continues to Recede over a Foot per Day , U.S. Army Corps of Engineers, New Orleans District Public Affairs .