Abstract
A better description of airborne transmission routes of viruses that are responsible for nosocomial infections requires efficient and accurate sampling methods that allow the preservation of viral integrity and infectivity. The aim of this project was to compare the virus collection efficiency of traditional filter sampling using cassettes with water-based condensation sampling through laminar flow. Bacteriophages MS2, PhiX174, Phi6, and PR772 as well as an influenza virus were nebulized in an aerosol chamber. Bioaerosols were simultaneously collected on polycarbonate (PC) filters (0.8 μm pore diameter) loaded into 37-mm closed-face cassettes (CFCs) and with a condensation-based sampler (Spot Sampler™) which collected bioaerosols in a liquid buffer. Concentrations of bacteriophages and influenza were analyzed at two different sampling times (60 min and 4 h) both by culture (infectivity preservation) and qPCR (efficiency of recovery) to compare the performance of the two samplers. PhiX174 showed better recovery with the Spot Sampler™ for both sampling times and PR 772 showed better relative genome recovery with CFC sampling after 60 min. In addition, use of the Spot Sampler™ led to better preservation of infectivity for all the viruses including influenza, with the exception of phage MS2 after 4 h of sampling. In future work, the Spot Sampler™ could be tested in hospital environments to better understand airborne transmission routes of viruses, or in in-vitro setups to assess the efficiency of virucidal air treatment.
Copyright © 2021 American Association for Aerosol Research
EDITOR:
1. Introduction
Recent respiratory viral outbreaks have become a worldwide concern. In 2018–2019 alone, 3,657 hospitalizations due to influenza were recorded in Canada (Public Health Agency of Canada Citation2019) and 496,600 in the USA (estimated death toll of 34,000). Most of these cases were the A/H1N1 strain (https://www.cdc.gov/flu/about/burden/2018-2019.html). In addition, the emergence of new outbreaks such as COVID-19 has caused new challenges in viral risk assessment. Understanding the transmission route of viruses is a challenge, but this knowledge could allow for much more efficient management of viral outbreaks. Airborne transmission of viruses such as influenza or SARS-CoV-2 has been recently investigated but needs further elucidation (Anderson et al. Citation2020; Fabian et al. Citation2008; Khedkar and Patzak Citation2020; Lei et al. Citation2018; Morawska and Cao Citation2020; Xiao et al. Citation2018). To evaluate the airborne spread of viruses, sampling methods with good physical collection efficiency and the ability to conserve the integrity of the sampled viruses are required. Without such methods, infectious viral concentrations could be underestimated due to sampling stress (Verreault, Moineau, and Duchaine Citation2008). Moreover, sensitivity to aerosolization and sampling conditions are specific to each strain of virus. Other environmental factors such as temperature, relative humidity, chemical composition of the air, and time spent in aerosol state also affect viral integrity in samples. The low concentration of viruses in the air, sensitivity of analytical methods, and collection efficiency are important aspects in the development of an aerosol characterization method for viruses (Hermann et al. Citation2006).
Liquid and dry impaction are the most common sampling methods used to study viral bioaerosol behavior, characteristics or destruction (Verreault, Moineau, and Duchaine Citation2008). The advantages and limitations of sampling using liquid impingers such as AGI-30 or AGI-4 (All Glass Impinger), SKC Biosampler®) and frit bubblers are well described (Hermann et al. Citation2006; Hogan et al. Citation2005; Hogan et al. Citation2006; Kettleson et al. Citation2009; Tseng and Li Citation2005; Walls et al. Citation2016). Collecting samples in liquid allows for better preservation of microorganisms and facilitates downstream analyses. A limitation of impingers for long-term sampling is the evaporation of liquid of collection and the re-aerosolization of biological particles, leading to the misestimation of the biological load (Lemieux et al. Citation2019).
The condensation of viral particles in a growth tube can also be used to study airborne viral particles (Hering and Stolzenburg Citation2005). Spot Sampler™ designed by Aerosol Devices Inc. applies this concept. This device uses water-based condensation of small particles achieved through laminar flow and that results in the gentle impaction of airborne particles. Particle condensation is performed in a growth tube with three distinct regions that are set at different temperatures: conditioner (inducing saturated vapor stream), initiator (supersaturation) and moderator (droplet growth by decreasing vapor content) (Hering, Spielman, and Lewis Citation2014; Hering and Stolzenburg Citation2005). Airborne particles can be collected in wet conditions in a vial containing a liquid (liquid Spot Sampler™), or in dry conditions in a 32-well plate (dry Spot Sampler™). Unlike impingers, which have a flow rate of 12.5 L/min, the Spot Sampler™ is a low-flow rate sampler (1.5 L/min).
Sampling by filtration using gelatin, polycarbonate (PC), or polytetrafluoroethylene (PTFE) filters are common in bioaerosol science for long-term sampling. The flow rate is usually set between 1 and 2 L/min, which is comparable to that of the Spot Sampler™. The type of filter used for these sampling methods can affect phage recovery. Previous studies using a similar aerosolization setup showed better recovery when using a PC filter than when using a PTFE filter for phage sampling (Gendron et al. Citation2010; Verreault et al. Citation2010).
To compare filtration versus water-based condensation sampling, total recovery (qPCR analysis) and infectivity (culture) were assessed using bacteriophages and a human virus.
Bacteriophages are often used as surrogates for pathogenic human viruses in environmental studies to assess viral particle transport, survival, the efficiency of protective equipment, and disinfection treatments (Agranovski et al. Citation2005; Dee et al. Citation2005; Grinshpun, Adhikari, and Honda Citation2007; Li et al. Citation2009; Tseng and Li Citation2006). All surrogates have limits and can produce biases in risk assessment, but these phages have the advantage of being safe for humans and easy to handle in laboratories (Sinclair et al. Citation2012). In the present study, four tailless bacteriophages were used. MS2 (family of Leviridae, single-stranded RNA) and Phi6 (Cystoviridae, double-stranded RNA) were used as surrogates for RNA viruses, and PR772 (Tectiviridae, double-stranded DNA) and PhiX174 (Microviridae, single-stranded DNA) were used as surrogates for DNA viruses (). In addition, a strain of influenza A (H1N1) (Orthomyxoviridae, single-stranded RNA) was used as a model for a human virus.
Table 1. Phages, viruses features and qPCR data for genome recovery.
Using bacteriophages and the influenza A strain, the present study aimed to compare the virus collection efficiency of the traditional sampling method using filters in cassettes and the sampling method using water-based condensation through laminar flow in a laboratory aerosol chamber.
2. Materials and methods
2.1. Set up
The setup was based on Verreault et al. (Citation2010) and is presented in . The figure shows where (1) air was injected in a (A) collection chamber (GenaMini chamber, SCL Meldtech Inc., Montreal, QC, Canada) where airflow rate generation was set at 3 L/min. The viruses were then aerosolized using an (B) atomizer (model 9302, TSI Inc., USA). The bioaerosols that were generated were then dried by being passed through a (C) desiccator (desiccants model 3062, TSI) before arriving back in the (A) GenaMini chamber. The dilution air rate was set at 2 L/min or at 7 L/min during APS measurements (Aerosol Particle Sizer model 3321, and Diluter model 3302 A, TSI Inc.). Bioaerosols were collected simultaneously on polycarbonate filters (0.8 μm pore diameter), loaded into (E) a 37-mm closed-face cassette (CFC) and the (D) Spot Sampler™. Flow rates were 2 L/min for CFCs (GilAir-5, Gilian, Sensidyne, LP, Clearwater, FL) and 1.5 L/min for the Spot Sampler™. Temperatures were set at 5 °C (conditioner), 40 °C (initiator), 21 °C (moderator), and 24 °C (nozzle) for sampling in liquid vials (liquid Spot Sampler™). Temperatures were set at 5 °C (conditioner), 35 °C (initiator), 10 °C (moderator), 28 °C (nozzle), and 35 °C (sample) for sampling in a well plate (dry Spot Sampler™). In addition, relative humidity and temperature were monitored ((F) Kimo RH 210) and bioaerosol stability was assessed by recording concentrations and size distribution (G). Results from APS measurements, relative humidity, and temperature for all sets of experiments are shown in . Excess air was collected in a (2) HEPA filter connected to a vacuum.
Figure 1. Experimental setup for aerosolization of bacteriophages and influenza in an aerosol chamber. The diagram is not to scale: (A) collection chamber, (B) atomizer, (C) desiccator, (D) Spot sampler™, (E) closed-face-cassette, (F) temperature/relative humidity probe, (G) Aerosol Particle Sizer, (1) air input (generation flowrate set 3 L/min in the collection chamber), and (2) air output.
Table 2. Environmental conditions and characteristics of bioaerosols (APS measurements) for each set of experiments.
2.2. Bacteriophage experiments
Phages and host strains were provided by the Félix d’Hérelle Reference Center for Bacterial Viruses (www.phage.ulaval.ca). Four bacteriophages were used as surrogates for eukaryotic viruses: Phi6 (HER 102) and MS2 (HER 462) for RNA phages, and PhiX174 (HER 036) and PR772 (HER 221) for DNA phages. The phages were amplified and cultured as described in (Turgeon et al. Citation2014). Phage concentrations were determined based on plaque assay analysis. Briefly, phages were incubated overnight at 37 °C in a TSA medium (Difco™ Tryptic Soy Agar, BD Biosciences, Canada) with their host strains: Escherichia coli HER 1036, HER 1221, and HER 1462 for PhiX174, PR772, and MS2, respectively. Phi6 and Pseudomonas syringae (HER1102) were incubated at 25 °C.
The phages were aerosolized in phage buffer composed of 20 mM of Tris-HCl (pH = 7.4), 100 mM of NaCl, and 10 mM of MgSO4 in sterile MilliQ water. A volume of 1 mL of each phage was added to the phage buffer for a final volume of 70 mL in the atomizer (TSI 9302 Atomizer). The initial phage concentration in the atomizer liquid was 4.10*1010 Plaque Forming Units (PFU)/mL for Phi6, 1.69*1010 PFU/mL for MS2, 1.63*108 PFU/mL for PhiX174, and 1.05*1010 PFU/mL for PR772. For all the experiments, 500 μL of the atomizer content was sampled before and after aerosolization for positive controls and to assess a potential loss of infectivity throughout the experimental timeline (no significant difference was observed).
Two types of experiments were conducted using phages: sampling in a vial of Spot Sampler™ liquid and sampling in a dry Spot sampler™ with phages collected in well plates. For all the experiments, phage suspensions were continuously aerosolized and sampled in CFCs and in the Spot Sampler™.
For Spot Sampler™ liquid sampling, two sampling times were tested (60 min and 4 h) in five independent assays. Aerosolization occurred the entire sampling period. Before each aerosolization, 200 μL of phage buffer was added in the vial. By the end of the aerosolization, the volume was completed to 350 μL for further analyses. For CFC sampling, 5 mL of phage buffer was added at the end of the aerosolization and after 60 min of agitation by orbital shaking (Boekel Ocelot Rotator, Fisher Scientific, USA), the liquid was eluted for further analyses.
Sampling using dry Spot Sampler™ was performed by collecting phage aerosols into wells for 60 min. After the 60-minute period, the plate switched automatically to the adjacent well. This was repeated five times (resulting in five wells per assay). Bioaerosols were simultaneously sampled in a CFC for the same duration. After 60 min of sampling, the CFC was moved to run under a laminar flow hood to simulate the same conditions as during well sampling. At the end of each assay, the five CFC samples were treated as described previously. A volume of 40 μL of preheated (37 °C) phage buffer was added to the wells three times to elute viral particles. Five independent sets were performed as described above (sets 1 to 5).
2.3. Influenza experiments
Influenza A/H1N1 (H1N1 wild type 2016 Michigan-like) was the human virus model. The virus was obtained by amplifying an initial stock solution in 10 ml of liquid medium composed of EMEM 1X (Wisent Inc., Canada), sodium bicarbonate (2.2 g/L, Wisent Inc., Canada), Hepes 1% (Wisent Inc., Canada) and a mix of Streptomycin/Penicillin 1% (Wisent Inc., Canada). The infection was performed by adding the viral suspension into a 150-cm2 flask containing MDCK (Madin-Darby Canine Kidney) cells after removing the culture medium and washing the cells with phosphate-buffered saline (PBS). After 60 min of incubation at 37 °C and 5% CO2, the medium was replaced by 20 mL of viral medium composed of EMEM 1X, 1% Hepes, and TPCK (Trypsin from bovine pancreas, Sigma-Aldrich, Canada). After 48 h of incubation at 37 °C and 5% CO2, MDCK cells were resuspended and the liquid was transferred into tubes to be centrifuged at 1,176 g for 10 min. Aliquots of the supernatant (30 mL for each tube) were stored at −80 °C until ready for use.
The first step for the influenza experiments was to compare infectivity preservation after 60 min by collecting bioaerosols or eluting particles in two different buffers, either in viral medium or in PBS 1 X (Lonza, USA). The contents from the two aliquots (60 mL) were put in the atomizer with 10 μL of Antifoam A Concentrate (Sigma-Aldrich, Canada) and aerosolized for 60 min in triplicate for each collection buffer. The second step was to aerosolize influenza for four hours (five independent essays). For this step, collection and particle elution were performed in PBS. Samples were collected using CFCs and the liquid Spot Sampler™ simultaneously, as previously described. Three aliquots were stored for both samplers (Spot Sampler™ and CFC) and the atomizer: 100 μL of liquid with 2 μL of BSA for culture assay, 150 μL of liquid for qPCR, and 100 μL of liquid with 2 μL of BSA (bovine albumin serum factor V, Gibco™, Canada) for backup. All samples were stored at −80 °C until analysis.
2.4. Genome recovery analysis by qPCR
Nucleic acids from RNA phages and influenza A were extracted using a QIAamp Viral RNA extraction Mini Kit (Qiagen, Germany). Phage RNA was converted into cDNA by RT-qPCR before being quantified by qPCR (). The master mix for retro transcription was composed of 4 μL of buffer, 1 μL of enzyme, 5 μL of RNA template, and 10 μL of water (iScript™ cDNA Synthesis Kit, BioRad). Temperature cycles were 25 °C for 5 min, 42 °C for 30 min, and 85 °C for 5 min using DNA Engine DYAD (Peltier thermal cycler, USA). The iQ supermix kit (BioRad, Canada) was used for qPCR for all phages. Influenza A recovery was assessed by one-step qPCR (iTaq™ universal probe one step kit, Biorad) (). For all the targeted sequences, amplification was performed using the CFX96™ Real-Time PCR Detection System (BioRad, Canada). To allow for quantification, standard curves were prepared using the method mentioned in several of the original studies (Gendron et al. Citation2010; Turgeon et al. Citation2014; Verreault et al. Citation2010). The results were expressed in copies/m3 for bioaerosols and copies/ml for atomizer content.
2.5. Calculations and statistical analyses
For the liquid Spot Sampler™ and CFC data comparison, the concentration (by plaque assay and qPCR) of phages in both air samples and the atomizer were calculated according to the methods previously described (Turgeon et al. Citation2014). The results were expressed in PFU or copies of genome/m3 for air samples, and PFU or copies of genomes/mL for the atomizer. To normalize data and to avoid bias due to initial concentrations of viruses in the atomizer, the relative recovery was calculated by dividing air sample concentrations by the initial atomizer content for each sampler. Finally, to assess the efficiency of the Spot Sampler™ compared to CFCs, relative recovery ratios for both devices were calculated for culture and qPCR results (Turgeon et al. Citation2014). Infectious influenza concentrations were determined by the TCID50 (Median Tissue Culture Infectious Dose), as described in a previous study (Reed and Muench Citation1938). All statistical tests were performed using Prism version 8.3 (GraphPad). To assess the culturability and genome recovery efficiency of the liquid Spot Sampler compared to CFCs, ratio-paired t-tests were performed on relative recovery ratios (Turgeon et al. Citation2014). The principle of this test is to calculate the logarithmic average of the treated ratios, as Spot Sampler/control (CFC). The null hypothesis (H0) is that the average of these ratios equals 0. A ratio equal to 1 means there is no difference between the treated sample compared to the control sample. The limit value to rejecting H0 was 0.05.
Concentrations measured with the dry Spot Sampler™ (and in associated CFCs) were normalized with atomizer concentrations in order to assess the effect of time on total recovery. The Mann-Whitney U test was performed to compare the efficiency of simultaneous sampling using the Spot Sampler™ and CFCs. Friedman’s test was used to assess the consistency of the measurements over time in the various wells of the dry Spot Sampler™.
3. Results
3.1. Liquid phage sampling
Detection limits by culture were 39 PFU/m3 for liquid Spot Sampler™ samples and 417 PFU/m3 for CFC samples. For molecular analysis, the detection limit varied according to the phage being examined but no significant difference of detection limit was observed for the liquid and dry Spot Sampler™ compared to the CFCs (data not shown). For both Spot Sampler, it was 7.7*103 copies/m3 of air for DNA phages and 1.6*104 copies/m3 for RNA phages.
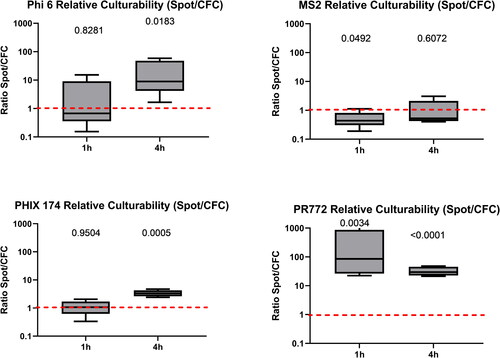
After 60 min of sampling, no difference in infectivity (culture) was observed for Phi6 and PhiX174, but sample collection using the liquid Spot Sampler™ allowed for a higher culture recovery of PR772 (p = 0.0034). However, the culturability of MS2 (p = 0.049) was better when CFCs () were used.
Figure 2. Ratio of relative culturability (Spot Sampler™/CFC) for all phages after 60 min and 4 h of sampling (n = 5 for each phage). A ratio greater than one indicates that the liquid Spot Sampler™ was more efficient at preserving the infectivity of phages. The value above each boxplot represents the p-value of the paired ratio t-test. A p-value below 0.05 means that the relative recovery between the Spot Sampler and CFC samples was statistically significantly different. There is one p-value for each sampling time. In the box plot, the whiskers represent the 5th and the 95th percentiles; the red line symbolizes a ratio of one, i.e., an eguality of relative culturability between the two samplers.
Genome recovery for PhiX174 (p = 0.0017) was higher when using the liquid Spot Sampler™, whereas CFC sampling led to better recovery of PR772 genomes (p = 0.0078) ().
Figure 3. Ratio of relative genome recovery (Spot Sampler™/CFC) for all phages after 60 min and 4 h of sampling (n = 5 for each phage). A ratio greater than one indicates that the liquid Spot Sampler™ was more efficient at recovering phage genomes. The value above each boxplot represents the p-value of the paired ratio t-test. A p-value below 0.05 means that the difference in relative recovery between Spot Sampler and CFC was statistically different. There is one p-value for each sampling time. In the box plot, the whiskers represent the 5th and the 95th percentiles; the red line symbolizes a ratio of one, i.e., an equality of genome recovery efficiency between the two samplers.
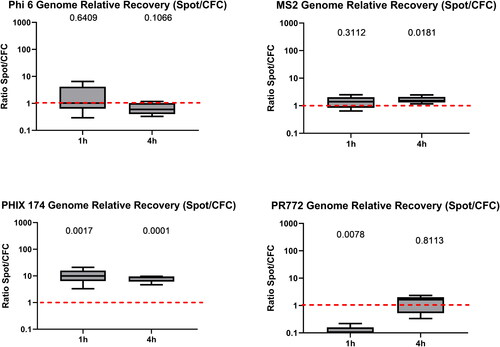
After 4 h of aerosolization, higher concentrations of all phages except MS2 were detected in liquid Spot Sampler™ samples by culture compared to CFC samples (). The median value for the preservation of infectivity was 9% (1.6% to 58%) higher in liquid Spot Sampler™ samples for Phi6, (2.4% to 4.7%) for PhiX174, and 30% (21% to48%) for PR772 after 4 h of aerosolization. No difference was observed for total recovery (qPCR analysis) with the exception of PhiX174 (p = 0.0001).
3.2. Phage sampling in dry condition
Dry well sampling (dry Spot Sampler™) did not affect total concentrations for Phi6 when compared to results seen from CFC sampling (). MS2 had a slight increase of less than one log for total recovery when using the dry Spot Sampler™ (Mann-Whitney: p = 0.035). For DNA bacteriophages (PhiX174 and PR772), total recovery was negatively affected using the dry Spot Sampler™, compared to CFCs (Mann-Whitney: p < 0.001).
Figure 4. Ratio of total recovery of genomes for four bacteriophages using the dry Spot Sampler™ and CFCs (n = 25 for each phage). The red line symbolizes a ratio of one, i.e., an equality of genome recovery efficiency between the two samplers.
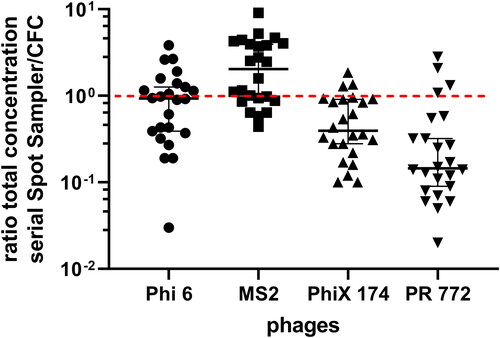
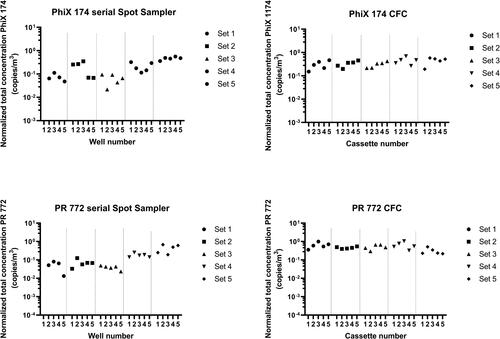
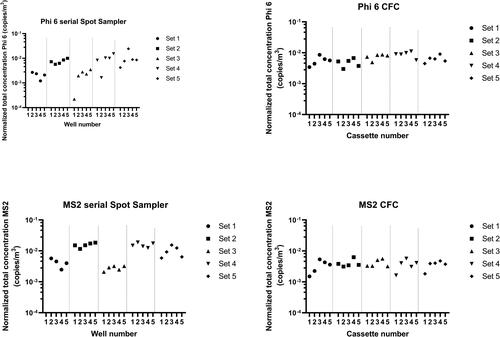
The homogeneity of recovery for samples collected in wells (dry Spot Sampler™) and in CFCs was assessed among sets for all phages ( and ). The variation of recovery was measured among sets of CFC samples for Phi6 with consistent results (Friedman: p = 0.0081), while the recovery of PhiX174 was inconsistent between CFC samples of the same set (Friedman: p = 0.048). However, differences in recoveries were observed using the dry Spot Sampler™ among sets for all bacteriophages (Friedman: Phi6 p = 0.0056, MS2 p = 0.0007, PhiX174 p = 0.004, and PR772 p = 0.007).
3.3. Influenza sampling in liquid
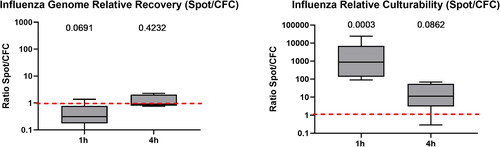
The collection buffer had no influence on the preservation of infectivity or on total recovery of influenza for both samplers (data not shown). The type of sampler used did not affect genome recovery regardless of the sampling duration (). After 60 min of sampling, the liquid Spot Sampler™ was better able to preserve culturability at a level that was three orders of magnitude higher than that of CFCs (p = 0.0003) (). After 4 h of sampling, no differences in total recovery and infectivity were observed between the two samplers.
Figure 7. Ratio of relative genome recovery and relative culturability (Spot Sampler™/CFC) for influenza after 60 min (n = 6) and 4 h (n = 5) of sampling. A ratio greater than one indicates that the liquid Spot Sampler™ was more efficient at preserving the genome and culturability of influenza. The value above each boxplot represents the p-value of the paired ratio t-test. A p-value below 0.05 means that the difference in relative recovery between the Spot Sampler™ and CFCs was statistically different. There is one p-value for each sampling time. In the box plot, the whiskers represent the 5th and the 95th percentiles; the red line symbolizes a ratio of one, i.e., an equality of genome recovery efficiency and of relative cuturability between the two samplers.
4. Discussion
The efficiency of these samplers was determined by several parameters such as airflow rate, collection principle, physical collection efficiency, and biological efficiency (genome integrity and infectivity preservation). The aim of this study was to assess the total recovery and preservation of infectivity of samples collected using the Spot Sampler™ and a PC filter in CFCs. The physical collection efficiency was not studied.
Previous studies have demonstrated that a water-based condensation sampler called the viable virus aerosol sampler (VIVAS) showed good physical efficiency for MS2 and influenza with 95% and over 90% efficiency respectively, while lower rates were observed for the SKC Biosampler®® (Lednicky et al. Citation2016; Pan et al. Citation2016). Indeed, another study showed that impingers such as the All Glass Impinger and SKC Biosampler® did not accurately collect submicrometer and ultrafine viral particles (Hogan et al. Citation2005).
In our study, the liquid Spot Sampler™ showed better preservation of infectivity for all phages after 4 h of sampling compared to the PC filter in CFCs. However, this was not the case for MS2, which is known to be a strong surrogate that is resistant to aerosolization (Trouwborst and De Jong Citation1973; Turgeon et al. Citation2014; Verreault et al. Citation2015; Walker and Ko Citation2007). PR772 was the only phage that showed better preservation of infectivity after 60 min of aerosolization, using liquid Spot sampler™. The type of sampler affected the total recovery for PR772, known to be sensitive to aerosolization (Turgeon et al. Citation2014). The other DNA phage PhiX174 was the only phage for which genome recovery was positively impacted by using the liquid Spot Sampler™, for both sampling times. Previous results suggest that liquid collection is a more appropriate sampling method than dry collection for PhiX174 (Turgeon et al. Citation2014). Other studies have reported improved collection efficiency for nanoscale particles by using water-based condensation samplers compared to impingers AGI-4 and SKC Biosampler® (Jiang et al. Citation2016; Walls et al. Citation2016).
The results for influenza were more contrasted with better preservation of infectivity after 60 min of sampling using the liquid Spot Sampler™ compared to CFC samples, but not after 4 h of sampling. The similarities between the values obtained for the two samplers that were observed for influenza after 4 h of sampling may be due to the large number of viruses in the filter leading to a protective effect by an agglomeration of viral particles, as described in previous studies (Galasso and Sharp Citation1965; Gerba and Betancourt Citation2017).
A previous study by Eiguren Fernandez and collaborators (Eiguren-Fernandez et al. Citation2014) reported the physical collection efficiency of the dry Spot Sampler™ to be 95% for a particulate size range of 0.010 to 2.5 μm. The authors also reported that for chemical compounds, there was no effect from the sampling time or storage duration in the sampler. In our study, we observed variations in phage concentrations among experiment sets. These concentrations were more constant for samples collected by filtration in CFCs, which confirms the reproducibility of the content of the aerosols that were produced during the different sets of experiments. The phage elution step after sampling with the serial Spot Sampler™ should be further optimized to improve the reproducibility of the results when using this device. It is also possible that viral particles were damaged during sampling, causing genome alteration.
The impact of filter type on the collection of viral particles has been well described in other reports (Gendron et al. Citation2010). However, there is no consensus on the best choice of filter for viral samples. PC filters were used because they have been shown to provide better preservation of infectivity compared to PTFE filters in previous studies using a similar aerosolization setup (Gendron et al. Citation2010; Verreault et al. Citation2010). Nevertheless, other studies have reported that PC filters induce stress due to dehydration and showed poor physical collection efficiency compared to PTFE or gelatin filters (Appert et al. Citation2012; Burton, Grinshpun, and Reponen Citation2006; Tseng and Li Citation2005).
Other parameters can affect the collection efficiency of virus samplers such as virus morphology and the nature of the virus (hydrophobic or hydrophilic viruses, DNA or RNA genome, etc.) (Tseng and Li Citation2005). Indeed, PR772 has been shown to be sensitive to aerosolization, similar to influenza (Agranovski et al. Citation2005; Turgeon et al. Citation2014; Verreault et al. Citation2015). Stress induced by aerosolization has also been documented to affect both genome integrity and infectivity, however, adding an organic fluid to the initial viral suspension can help preserve viral particle integrity (Turgeon et al. Citation2014). The addition of salts in low concentrations can also improve the preservation of viral particles, but the effects are varied depending on the virus (Benbough Citation1971; Trouwborst and De Jong Citation1973; Walker and Ko Citation2007). A study by Pyankov at al. (Pyankov, Pyankova, and Agranovski Citation2012) reported that the strain of influenza could affect its preservation of infectivity in ambient air. Moreover, the size of viral particles affects physical and biological collection efficiency (Hogan et al. Citation2005; Zuo et al. Citation2014; Zuo et al. Citation2013). Despite these findings, the morphology and structure of the virus alone cannot predict aerosolization resistance in viral particles (Ijaz et al. Citation1985).
The duration of sampling is another factor that can also affect the assessment of viral exposure (Hogan et al. Citation2005). Phage Phi6 exhibited signs of significant damage as a result of 20 min of sampling by filtration using PC and PTFE filters (Gendron et al. Citation2010). However, this was not observed in our study, where all phages and the influenza strain were detected using both samplers. The liquid Spot Sampler™ provided results that indicated a good ability to preserve the infectivity of influenza and phages for long-term sampling.
Temperature and relative humidity parameters must be considered when measuring airborne viruses. In the present study, the temperature and relative humidity reflected normal ambient air, with temperatures between 20 °C and 25 °C and relative humidity ranging from 20% to 30%. However, the optimal conditions needed to preserve phages and viruses vary according to the surrogate (Harper Citation1961). Some studies have reported that MS2 sampling was either not affected by environmental conditions or was better at low or average relative humidity (RH) (Appert et al. Citation2012; Trouwborst and De Jong Citation1973). However, another study showed PhiX174 to be better preserved at a high RH (80%) and PR772 to be sensitive to aerosolization at all RHs (Appert et al. Citation2012; Trouwborst and De Jong Citation1973; Verreault et al. Citation2015). It seems that most influenza strains remain stable at low RH levels. Relative humidity can also affect viral particles size distribution during long periods of aerosolization (Harper Citation1961; Wang and Brion Citation2007).
5. Conclusion
Using the liquid Spot Sampler™ for sample collection resulted in better preservation of phage and influenza infectivity for long sampling periods when compared to PC filters in CFCs. Spot Samplers™ only require a small volume (up to 400 μL) of liquid to elute (dry Spot Sampler™) or collect (liquid Spot Sampler™) viral particles, compared to the amount necessary for impingers or filters (several ml depending on the sampler). An advantage of this device is its ability to concentrate viruses, which increases the probability of detecting viral particles that are otherwise often present in low concentrations in the environment. The dry Spot Sampler™ could be a useful tool for monitoring over long periods after particle elution has been optimized. In general, devices that use water-based condensation as a sample collection strategy seems to be relevant for the assessment of airborne viral exposure. The positive results for the preservation of infectivity make the liquid Spot Sampler a useful tool to assess the efficiency of various disinfection methods.
In future work, the liquid Spot Sampler™ should be tested on the field while ensuring the disinfection of the device after contact with a pathogen.
Acknowledgment
The authors would like to thank Vincent Brochu for his technical support, Dr Guy Boivin’s laboratory for providing guidance and cells for the influenza culture, and the Réseau en Santé Respiratoire du Québec for the post-doctoral fellowship (JD). This work was supported by the National Sciences and Engineering Research Council (NSERC) Discovery grant (CD). CD holds the Tier-1 Canada Research Chair on Bioaerosols.
Conflicts of interest
The authors declare no conflicts of interest and have not received any financial support from any of the private companies mentioned.
Additional information
Funding
References
- Agranovski, I. E., A. S. Safatov, O. V. Pyankov, A. A. Sergeev, A. N. Sergeev, and S. A. Grinshpun. 2005. Long-term sampling of viable airborne viruses. Aerosol Sci. Technol. 39 (9):912–8. doi:https://doi.org/10.1080/02786820500297012.
- Anderson, E. L., P. Turnham, J. R. Griffin, and C. C. Clarke. 2020. Consideration of the aerosol transmission for covid-19 and public health. Risk Anal. 40 (5):902–7. doi:https://doi.org/10.1111/risa.13500.
- Appert, J., P. C. Raynor, M. Abin, Y. Chander, H. Guarino, S. M. Goyal, Z. Zuo, S. Ge, and T. H. Kuehn. 2012. Influence of suspending liquid, impactor type, and substrate on size-selective sampling of ms2 and adenovirus aerosols. Aerosol Sci. Technol. 46 (3):249–57. doi:https://doi.org/10.1080/02786826.2011.619224.
- Benbough, J. E. 1971. Some factors affecting the survival of airborne viruses. J. Gen. Virol. 10 (3):209–20. doi:https://doi.org/10.1099/0022-1317-10-3-209.
- Burton, N. C., S. A. Grinshpun, and T. Reponen. 2006. Physical collection efficiency of filter materials for bacteria and viruses. Ann. Occup. Hyg. 51:143–51. doi:https://doi.org/10.1093/annhyg/mel073.
- Dee, S. A., J. Deen, L. Jacobson, K. D. Rossow, C. Mahlum, and C. Pijoan. 2005. Laboratory model to evaluate the role of aerosols in the transport of porcine reproductive and respiratory syndrome virus. Vet. Rec. 156:501–4. doi:https://doi.org/10.1136/vr.156.16.501.
- Eiguren-Fernandez, A., G. S. Lewis, S. R. Spielman, and S. V. Hering. 2014. Time-resolved characterization of particle associated polycyclic aromatic hydrocarbons using a newly-developed sequential spot sampler with automated extraction and analysis. Atmos Environ (1994) 96:125–34. doi:https://doi.org/10.1016/j.atmosenv.2014.07.031.
- Fabian, P., J. J. McDevitt, W. H. DeHaan, R. O. P. Fung, B. J. Cowling, K. H. Chan, G. M. Leung, and D. K. Milton. 2008. Influenza virus in human exhaled breath: An observational study (influenza virus in breath). PLoS ONE 3 (7):e2691. doi:https://doi.org/10.1371/journal.pone.0002691.
- Galasso, G. J., and D. G. Sharp. 1965. Effect of particle aggregation on the survival of irradiated vaccinia virus. J. Bacteriol. 90 (4):1138–42. doi:https://doi.org/10.1128/JB.90.4.1138-1142.1965.
- Gendron, L., D. Verreault, M. Veillette, S. Moineau, and C. Duchaine. 2010. Evaluation of filters for the sampling and quantification of rna phage aerosols. Aerosol Sci. Technol. 44 (10):893–901. doi:https://doi.org/10.1080/02786826.2010.501351.
- Gerba, C. P., and W. Q. Betancourt. 2017. Viral aggregation: impact on virus behavior in the environment. Environ. Sci. Technol. 51 (13):7318–25. doi:https://doi.org/10.1021/acs.est.6b05835.
- Grinshpun, S. A., A. Adhikari, and T. Honda. 2007. Control of aerosol contaminants in indoor air: Combining the particle concentration reduction with microbial inactivation. Environ. Sci. Technol. 41 (2):606–12. doi:https://doi.org/10.1021/es061373o.
- Harper, G. J. 1961. Airborne micro-organisms: Survival tests with four viruses. J. Hyg. (Lond). 59:479–86. doi:https://doi.org/10.1017/S0022172400039176.
- Hering, S. V., S. R. Spielman, and G. S. Lewis. 2014. Moderated, water-based, condensational particle growth in a laminar flow. Aerosol Sci. Technol. 48 (4):401–8. doi:https://doi.org/10.1080/02786826.2014.881460.
- Hering, S. V., and M. R. Stolzenburg. 2005. A method for particle size amplification by water condensation in a laminar, thermally diffusive flow. Aerosol Sci. Technol. 39 (5):428–36. doi:https://doi.org/10.1080/027868290953416.
- Hermann, J. R., S. J. Hoff, K. J. Yoon, A. C. Burkhardt, R. B. Evans, and J. J. Zimmerman. 2006. Optimization of a sampling system for recovery and detection of airborne porcine reproductive and respiratory syndrome virus and swine influenza virus. Appl. Environ. Microbiol. 72 (7):4811–8. doi:https://doi.org/10.1128/AEM.00472-06.
- Hogan, C. J., E. M. Kettleson, M. H. Lee, B. Ramaswami, L. T. Angenent, and P. Biswas. 2005. Sampling methodologies and dosage assessment techniques for submicrometre and ultrafine virus aerosol particles. J. Appl. Microbiol. 99 (6):1422–34. doi:https://doi.org/10.1111/j.1365-2672.2005.02720.x.
- Hogan, C. J., E. M. Kettleson, B. Ramaswami, D.-R. Chen, and P. Biswas. 2006. Charge reduced electrospray size spectrometry of mega- and gigadalton complexes: Whole viruses and virus fragments. Anal. Chem. 78 (3):844–52. doi:https://doi.org/10.1021/ac051571i.
- Ijaz, M. K., A. H. Brunner, S. A. Sattar, R. C. Nair, and C. M. Johnson-Lussenburg. 1985. Survival characteristics of airborne human coronavirus 229e. J Gen Virol. 66 (12):2743–8. doi:https://doi.org/10.1099/0022-1317-66-12-2743.
- Jiang, X., M. Pan, S. V. Hering, J. A. Lednicky, C.-Y. Wu, and Z. H. Fan. 2016. Use of rna amplification and electrophoresis for studying virus aerosol collection efficiency and their comparison with plaque assays. Electrophoresis. 37 (19):2574–80. doi:https://doi.org/10.1002/elps.201600141.
- Kettleson, E., B. Ramaswami, C. J. Hogan, M. Lee, G. Statyukha, P. Biswas, and L. Angenent. 2009. Airborne virus capture and inactivation by an electrostatic particle collector. Environ. Sci. Technol. 43 (15):5940–6. doi:https://doi.org/10.1021/es803289w.
- Khedkar, P. H., and A. Patzak. 2020. Sars-cov-2: What do we know so far? Acta. Physiol. (Oxf). 229 (2):e13470 doi:https://doi.org/10.1111/apha.13470.
- Lednicky, J., M. Pan, J. Loeb, H. Hsieh, A. Eiguren-Fernandez, S. Hering, Z. H. Fan, and C.-Y. Wu. 2016. Highly efficient collection of infectious pandemic influenza h1n1 virus (2009) through laminar-flow water based condensation. Aerosol Sci. Technol. 50 (7):i–iv. doi:https://doi.org/10.1080/02786826.2016.1179254.
- Lei, H., Y. Li, S. Xiao, C. H. Lin, S. L. Norris, D. Wei, Z. Hu, and S. Ji. 2018. Routes of transmission of influenza a h1n1, sars cov, and norovirus in air cabin: Comparative analyses. Indoor Air. 28 (3):394–403. doi:https://doi.org/10.1111/ina.12445.
- Lemieux, J., M. Veillette, H. Mbareche, and C. Duchaine. 2019. Re-aerosolization in liquid-based air samplers induces bias in bacterial diversity. Aerosol Sci. Technol. 53 (11):1244–60. doi:https://doi.org/10.1080/02786826.2019.1652242.
- Li, H.-W., C.-Y. Wu, F. Tepper, J.-H. Lee, and C. N. Lee. 2009. Removal and retention of viral aerosols by a novel alumina nanofiber filter. J. Aerosol Sci. 40 (1):65–71. doi:https://doi.org/10.1016/j.jaerosci.2008.09.003.
- Morawska, L., and J. Cao. 2020. Airborne transmission of sars-cov-2: The world should face the reality. Environ. Int. 139:105730 doi:https://doi.org/10.1016/j.envint.2020.105730.
- Pan, M., A. Eiguren-Fernandez, H. Hsieh, N. Afshar-Mohajer, S. V. Hering, J. Lednicky, Z. Hugh Fan, and C.-Y. Wu. 2016. Efficient collection of viable virus aerosol through laminar-flow, water-based condensational particle growth. J. Appl. Microbiol. 120 (3):805–15. doi:https://doi.org/10.1111/jam.13051.
- Public Health Agency of Canada. 2019. Fluwatch: 2018-2019 influenza season.
- Pyankov, O. V., O. G. Pyankova, and I. E. Agranovski. 2012. Inactivation of airborne influenza virus in the ambient air. J. Aerosol Sci. 53:21–8. doi:https://doi.org/10.1016/j.jaerosci.2012.05.011.
- Reed, L. J., and H. Muench. 1938. A simple method of estimating fifty per cent endpoints. Am. J. Epidemiol. 27 (3):493–7. doi:https://doi.org/10.1093/oxfordjournals.aje.a118408.
- Sinclair, R. G., J. B. Rose, S. A. Hashsham, C. P. Gerba, and C. N. Haas. 2012. Criteria for selection of surrogates used to study the fate and control of pathogens in the environment. Appl. Environ. Microbiol. 78 (6):1969–77. doi:https://doi.org/10.1128/AEM.06582-11.
- Trouwborst, T., and J. C. De Jong. 1973. Interaction of some factors in the mechanism of inactivation of bacteriophage ms2 in aerosols. Appl. Microbiol. 26 (3):252–7. doi:https://doi.org/10.1128/AEM.26.3.252-257.1973.
- Tseng, C.-C., and C.-S. Li. 2005. Collection efficiencies of aerosol samplers for virus-containing aerosols. J. Aerosol Sci. 36 (5):593–607. doi:https://doi.org/10.1016/j.jaerosci.2004.12.004.
- Tseng, C.-C., and C.-S. Li. 2006. Ozone for inactivation of aerosolized bacteriophages. Aerosol Sci. Technol. 40 (9):683–9. doi:https://doi.org/10.1080/02786820600796590.
- Turgeon, N., M.-J. Toulouse, B. Martel, S. Moineau, and C. Duchaine. 2014. Comparison of five bacteriophages as models for viral aerosol studies. Appl. Environ. Microbiol. 80 (14):4242–50. doi:https://doi.org/10.1128/AEM.00767-14.
- Verreault, D., M. Marcoux-Voiselle, N. Turgeon, S. Moineau, and C. Duchaine. 2015. Resistance of aerosolized bacterial viruses to relative humidity and temperature. Appl. Environ. Microbiol. 81 (20):7305–11. doi:https://doi.org/10.1128/AEM.02484-15.
- Verreault, D., S. Moineau, and C. Duchaine. 2008. Methods for sampling of airborne viruses. Microbiol. Mol. Biol. Rev. 72 (3):413–44. doi:https://doi.org/10.1128/MMBR.00002-08.
- Verreault, D., G. M. Rousseau, L. Gendron, D. Massé, S. Moineau, and C. Duchaine. 2010. Comparison of polycarbonate and polytetrafluoroethylene filters for sampling of airborne bacteriophages. Aerosol Sci. Technol. 44 (3):197–201. doi:https://doi.org/10.1080/02786820903518899.
- Walker, C., and G. Ko. 2007. Effect of ultraviolet germicidal irradiation on viral aerosols. Environ. Sci. Technol. 41 (15):5460–5. doi:https://doi.org/10.1021/es070056u.
- Walls, H. J., D. S. Ensor, L. A. Harvey, J. H. Kim, R. T. Chartier, S. V. Hering, S. R. Spielman, and G. S. Lewis. 2016. Generation and sampling of nanoscale infectious viral aerosols. Aerosol Sci. Technol. 50 (8):802–11. doi:https://doi.org/10.1080/02786826.2016.1191617.
- Wang, M., and G. Brion. 2007. Effects of rh on glass microfiber filtration efficiency for airborne bacteria and bacteriophage over time. Aerosol Sci. Technol. 41 (8):775–85. doi:https://doi.org/10.1080/02786820701455351.
- Word Health Organization. 2009. Cdc protocol of realtime rtpcr for swine influenza a(h1n1). https://www.who.int/csr/resources/publications/swineflu/CDCrealtimeRTPCR protocol_20090428.pdf?fbclid=IwAR1zah_yETUv5W-z0Ju T07HyldG0g9a0Y4DeCdRxqX8tTzNo3OSOeY9l0HA.
- Xiao, S., J. W. Tang, D. S. Hui, H. Lei, H. Yu, and Y. Li. 2018. Probable transmission routes of the influenza virus in a nosocomial outbreak. Epidemiol. Infect. 146 (9):1114–22. doi:https://doi.org/10.1017/S0950268818001012.
- Zuo, Z.,. T. H. Kuehn, A. Z. Bekele, S. K. Mor, H. Verma, S. M. Goyal, P. C. Raynor, and D. Y. H. Pui. 2014. Survival of airborne ms2 bacteriophage generated from human saliva, artificial saliva, and cell culture medium. Appl. Environ. Microbiol. 80 (9):2796–803. doi:https://doi.org/10.1128/AEM.00056-14.
- Zuo, Z.,. T. H. Kuehn, H. Verma, S. Kumar, S. M. Goyal, J. Appert, P. C. Raynor, S. Ge, and D. Y. H. Pui. 2013. Association of airborne virus infectivity and survivability with its carrier particle size. Aerosol Sci. Technol. 47 (4):373–82. doi:https://doi.org/10.1080/02786826.2012.754841.