Abstract
Mutations in the TP53 gene are a well-documented strong prognostic factor in breast cancer. A prognostic value of the Arg72Pro polymorphism of the TP53 gene is more contradictory. We assessed TP53 mutations and genotypes of the Arg72Pro polymorphism in a study including 204 Danish women. Patients with mutations in the TP53 gene had a significant reduction in disease-free survival of breast cancer (p < 0.0001). Genotypes of the Arg72Pro polymorphism were neither significantly associated with TP53 mutations nor with disease-free survival (p = 0.4). Among heterozygous patients a reduction in disease-free survival was found for patients with LOH and retention of the Pro allele as compared to patients with LOH and retention of the Arg allele and patients with no LOH (p = 0.05). In conclusion, we find a highly significant prognostic value of TP53 mutations but find a possible prognostic value of the Arg72Pro polymorphism only related to LOH.
Numerous studies have shown that mutations in the TP53 gene (obtained using sequencing of tumor DNA) represent a strong prognostic factor in breast cancer. In fact, several of the studies, including a study by our group, showed that the presence of a TP53 mutation is the single most adverse prognostic indicator of both recurrence of disease and death Citation[1–3].
Codon 72 of the TP53 gene harbors a well-known polymorphic site with the two alleles arginine (Arg) and proline (Pro) (Arg72Pro). The site is located in a hydrophobic region of the TP53 gene (between amino acids 61 and 94) Citation[4]. The region contains five repeats of the sequence PXXP (in which P represents Pro and X represents any amino acid) with codon 72 participating in one of them. Deletion of this region was shown to impair the ability of the p53 protein to suppress tumor cell growth in culture Citation[5]. In addition the p53 protein containing proline at codon 72 was shown to be structurally different from the p53 protein containing arginine, reflected by its altered electrophoretic mobility Citation[6]. The Arg72Pro polymorphism in exon 4 has been related to risk of breast cancer development as well as survival from breast cancer. Risk of breast cancer development has been associated with patients carrying Arg/Arg Citation[7], Citation[8], with patients carrying Pro/Pro Citation[9], Citation[10] and with no correlation to the Arg72Pro polymorphism Citation[11–13]. Diverging results also exist regarding a potential prognostic value of the polymorphism Citation[13–17]. Tommiska et al. found in an unselected group of 888 breast cancer patients a significantly poorer survival for patients homozygous for the Pro allele as compared to patients homozygous for the Arg allele or heterozygous. Being homozygous for the Pro allele was an independent prognostic factor in multivariate analyses and it was independent of p53 protein overexpression. It should, however, be emphasized that p53 protein overexpression is not as strong a prognostic marker as is TP53 mutations obtained by sequencing Citation[1] and a poor correlation between p53 protein overexpression and nonsense mutations and deletions has been reported Citation[18], Citation[19]. A recent study by Xu et al. also reported a poorer pathological response to anthracycline-based neoadjuvant chemotherapy for patients homozygous for the codon 72 Pro allele Citation[20]. Goode et al. Citation[16] reported in contrast that improved breast cancer survival was associated with patients carrying the Pro allele as shown in a large population-based study of more than 2 000 patients. It was concluded, however, that the genotype was unlikely to replace current markers of prognosis such as the estrogen receptor status, and besides they did not stratify for TP53 mutations Citation[16]. Bonafe et al. reported that in heterozygous tumors with codon 72 loss of heterozygosity (LOH), retention of the Arg allele was associated with reduced disease-free and overall survival Citation[17]. They, as well, did not examine TP53 mutations.
In conclusion, diverging results have been reported regarding the Arg72Pro polymorphism and risk of breast cancer development as well as correlation with breast cancer prognosis. As we have found no breast cancer studies examining TP53 mutations, TP53 codon 72 genotypes, LOH, and specific allele retention together, we decided to examine all four parameters in this study of 204 Danish breast cancer patients.
Materials and methods
Patients
Our patient group is part of a large consecutive group of 455 patient previously described in detail Citation[21]. The 455 patients were diagnosed with primary breast carcinoma in the period from January 1990 to 1994 and fulfilled the following inclusion criteria: having primary unilateral breast carcinoma with no evidence of disseminated disease; availability of complete clinical and histopathological information; having no other malignancies; having received radical therapy. Additional inclusion criteria for the large group of patients were: the patients had their tumor biopsies submitted for estrogen receptor analysis and tumor material was available for further studies. The study was carried out with approval for the ethics committee.
This study comprised 204 out of 455 patients characterized by having available DNA extracted from blood samples. The patients were followed to death or to July 1, 2004, which gives a median potential observation time of 32 months for the patients that died before July 1, 2004 and of 120 months for the patients alive onJuly 1, 2004.
Treatment
Locoregional treatment was either lumpectomy or total mastectomy, both with axillary lymph node dissection including removal of the central axillary lymph nodes involving level I and II. All patients treated with lumpectomy were given adjuvant radiotherapy (48 Gy/24 fx., 5 fx. weekly) to residual breast and women younger than 45 years with >3 positive lymph nodes also had their regional lymph nodes irradiated. Mastectomized patients younger than 45 years of age with more than three positive lymph nodes received adjuvant radiotherapy to the thorax wall and regional lymph nodes. Patients had systemic therapy according to the national Danish treatment policy described by The Danish Breast Cancer Cooperation group (DBCG 89 protocols). Patients were treated according to risk status, estrogen receptor status and menopausal status including CMF or CEF plus/minus Pamindronate to high-risk premenopausal women and to high-risk postmenopausal women, Tamoxifen for different periods of time. This has previously been described in detail Citation[21].
Tumor and normal DNA
DNA was extracted from purified tumor cells and blood lymphocytes. The extraction procedures has been described in detail in previous publications Citation[22], Citation[23].
Assessment of TP53 mutations in tumor tissue
TP53 mutations have previously been assessed by denaturing gradient gel electrophoresis (DGGE) analysis and DNA sequencing in this cohort. The mutations were separated in different groups according to function of the affected amino acids as previously described in details Citation[2], Citation[3].
Assessment of the Arg72Pro polymorphism and LOH
PCR amplification
The TP53 codon 72 alleles were amplified with AmpliTaq GOLD polymerase (Applied Biosystems, Foster City, CA, USA) using the primer pairs 5′-ACC TGG TCC TCT GAC TGC-3′ and 5′-GCC GCC GGT GAA AAT AGG AGC TG-3′. Amplification was performed under the following conditions: initial denaturation: 95°C for 12 min, followed by 35 rounds of thermal cycling (95°C for 30 s, 58°C for 30 s and 72°C for 30 s) and a final extension at 72°C for 10 min. The PCR products were separated through a 1.25% agarose gel containing 0.5 µg/ml ethidium bromide, the DNA bands were located by UV light, cut and excised from the gels and filtered by a Montage DNA Gel Extraction Device (5.000 g/10 min), Millipore Corporation, Bedford, MA, USA.
Primer extension
Primer extension reaction was performed according to a recently described method Citation[24]. Primers were designed allowing extension of a 25 base pair product when the base at codon 72 was a guanine (G) and extension of a 30 base pair product when the base was a cytosine (C). The primer extension reaction was performed in a final reaction volume of 20 µl containing: 0.3 µM of the extension primer sequence 5′-AGG AGC TGC TGG TGC AGG GGC CAC G-3′, 50 µM of respectively ddATP, ddCTP and dGTP, 0.96 Unit of Thermo Sequenase (Amersham), 4 µl of purified PCR product and 1×reaction buffer. The primer extension reaction was performed under the following conditions: 96°C for 1 min, 50 cycles of 96°C for 10 s, 50°C for 15 s and 60°C for 1 min, and a final denaturation of 96°C for 30 s Citation[24]. Denaturing high performance liquid chromatography (DHPLC) analysis was conducted on the automated Wave DNA fragment analysis system (Transgenomic, Omaha, NE, USA). Twelve µl of primer extension reaction from each patient sample was injected into the mobile phase (Buffer A 0.1 mol/l triethylamine acetate (TEAA); Buffer B 0.1 mol/l TEAA/25% acetonitrile) at 0.9 ml/min flow rate. The primer extension products were eluted from the solid phase (DNASep Cartridge; Transgenomic, Omaha, NE, USA) by a linear gradient (18–39% buffer B) in a 9-min sample-run under fully denaturing conditions (70°C). The eluted products were mixed with an intercalating dye (DNA Staining Solution I, Transgenomic, Omaha, NE, USA), detected by fluorescence and analyzed using Transgenomic Navigator Software.
Arg72Pro study design
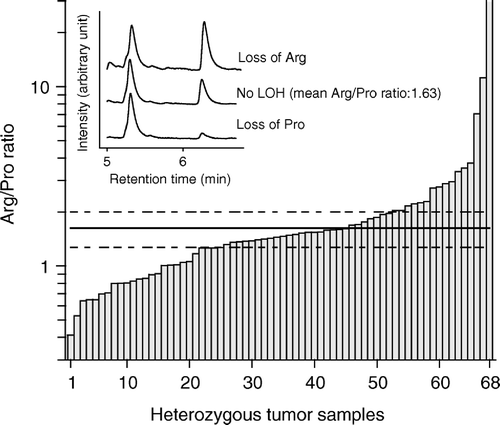
shows the elution profiles of three representative tumor samples. A peak between 5.2 to 5.5 minutes denotes an arginine allele and a peak between 6.2 to 6.5 minutes denotes a proline allele. The TP53 codon 72 genotype was assessed on DNA from normal tissue for all 204 patients and the analysis was repeated for the 69 heterozygous patients. For the heterozygous patients two ratios, obtained dividing the area under the arginine peak with the area under the proline peak, were compiled and a mean was calculated for each of the heterozygous patients. Plotting the mean Arg/Pro ratios of the heterozygous normal tissue samples in a histogram and in a Q-Q plot demonstrated a normal distribution and a significance level of 99% were calculated. Tumor DNA was analyzed in triplicates and the mean Arg/Pro ratio was calculated for 68 of the heterozygous tumor samples (no result was obtained for one sample). A tumor sample was categorized with LOH if the mean Arg/Pro ratio was outside the borders indicating a 99% significance level of the normal tissue samples. (main figure) shows the mean ratios of each heterozygous tumor sample. In addition, lines representing the 99% significance borders of the normal tissue samples are included. To validate our method we performed a standard RFLP Citation[25] on some of the samples homozygous for Arg or Pro and heterozygous with or without LOH (data not shown).
Statistical evaluation
Associations between categorical data were tested either by two-sided χ2 test or by a non parametric exact test. Disease-free survival functions were made according to the Kaplan-Meier method and the differences between the curves were calculated according to the log-rank test. Associations between individual clinico-pathological variables and disease-free survival were assessed using the Cox proportional hazard regression model. The level of statistical significance was 5% and p-values were estimated by a two-tailed test. The analyses were conducted using STATA 8.2 statistical software.
Results
Clinico-pathological parameters and TP53 mutations
Seven patients out of 204 patients developed an isolated loco-regional recurrence during the follow-up, 29 patients had both loco-regional recurrence and distant metastases, 49 patients developed distant metastases, and a total of 67 patients died of breast cancer and another 20 patients died from other reasons. is a presentation of the 8-year actuarial probability of disease-free survival, for known classical clinico-pathological parameters. TP53 mutations and the Nottingham Prognostic Index, which is based on tumor size, tumor grade, and axillary lymph node status Citation[26], are included in the table. Increasing number of positive lymph nodes, increasing risk according to the Nottingham Prognostic Index, aggressive or very aggressive TP53 mutations, and large tumor size are important indicators of recurrence and mortality. In a multivariate Cox proportionate regression analysis TP53 mutations and positive lymph nodes were the only parameters remaining significant at a 5% significance level. Similar results were obtained using another classification of TP53 mutations based on whether or not the mutation affected the L2 and L3 domains Citation[19] (data not shown).
Table I. Eight years’ actuarial probability of disease-free survival in 204 patients diagnosed with breast cancer
TP53 codon 72 genotypes
The allele frequencies of the Arg72Pro polymorphism among the 204 breast cancer patients were 0.74 and 0.26 for the Arg72 and Pro72 allele, respectively, and the polymorphism was shown to be in Hardy-Weinberg equilibrium. The exact same allele frequencies (0.74 and 0.26) have previously been reported in Danish controls Citation[27]. Of the 204 breast cancer patients 116 (57%) carried Arg/Arg, 19 (9%) carried Pro/Pro and 69 (34%) carried Arg/Pro. We discovered no significant differences in the distribution of the three genotypes focusing on clinico-pathological parameters ().
Table II. Distribution of clinicopathological and biopathological parameters in breast cancer patients with different genotypes of TP53 codon 72
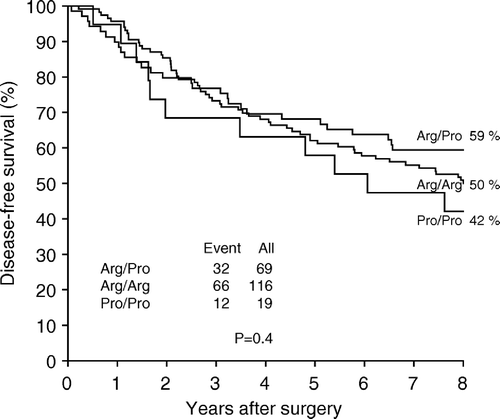
Of the 116 patients carrying Arg/Arg, 23 patients (20%) also carried an aggressive or very aggressive tumor specific TP53 mutation. Of the 19 patients carrying Pro/Pro 5 patients (26%) carried a TP53 mutation. This difference in distribution of TP53 mutations was not statistically significant (p > 0.2). We stratified our data according to the Nottingham Prognostic Index (NPI). Among the 25 patients with the poorest prognosis (NPI > 5.4) 6 of 13 patients (46%) carrying Arg/Arg at codon 72 and 1 of 4 patients (25%) carrying Pro/Pro also carried a TP53 mutation. Among the 179 patients in the good/intermediate prognostic group (NPI < 5.4) 17 of 103 patients (17%) carrying Arg/Arg and 4 of 15 patients (27%) carrying Pro/Pro also carried a TP53 mutation. However, the finding was not statistically significant (p = 0.7). The three different codon 72 genotypes: Arg/Arg, Pro/Pro and Arg/Pro did not differ significantly with respect to disease-free survival (p = 0.4) (). Stratifying the patients according to TP53 mutations in a group with TP53 wt or TP53 neutral mutations and in a group with TP53 aggressive mutations neither recovered a difference in survival for the codon 72 genotypes.
Tumor-LOH of TP53 codon 72
Twenty eight of 69 patients heterozygous of codon 72 were inside the upper and lower control lines (99% significance borders of the normal tissue) indicating no tumor-LOH and 40 of 69 patients (58%) were found outside the control lines indicating tumor-LOH. No PCR product was obtained from one tumor.
Sixteen of 40 patients (40%) with LOH carried relatively more of the Arg allele as compared to the Pro allele (referred to as retention of Arg) and 24 patients (60%) carried relatively more of the Pro allele as compared to the Arg allele (referred to as retention of Pro).
The distribution of clinico-pathological parameters among heterozygous patients with LOH and retention of respectively Arg or Pro and heterozygous patients with no LOH did not differ significantly, except for malignancy grade and TP53 mutations ().
Table III. Distribution of clinicopathological and biopathological parameters in heterozygous breast cancer patients with LOH and retention of either Arg or Pro or with no LOH of TP53 codon 72
Only 1 of 28 patients with no LOH carried a TP53 mutation (a nonsense mutation, Q331X, within the C-terminal tetramerization domain) as compared to 10 of 40 patients with LOH (5 patients with retention of Arg and 5 patients with retention of Pro) (p = 0.04).
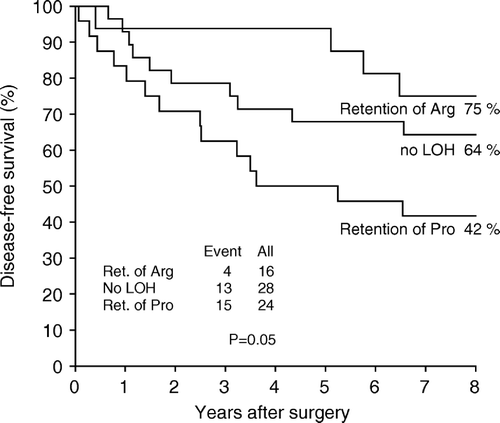
No significant reduction in disease-free survival was found for heterozygous patients with LOH as compared to no LOH (p > 0.2) However, separating the group of patients with LOH according to specific allele retention, a significant reduction in disease-free survival was found for the group of patients with retention of the Pro allele as compared to the group of patients with retention of the Arg allele and the group of patients with no LOH (p = 0.05) (). We stratified the group of heterozygous patients according to TP53 mutations. In the group accounting 11 patients with TP53 mutations we found a significant reduction in disease-free survival for patients with retention of the Pro allele as compared to patients with retention of the Arg allele and patients with no LOH (P = 0.04). In the group of 57 patients with TP53 wild type or neutral mutations we did not, however, find a statistically significant reduction in survival for patients with retention of the Pro allele (p = 0.3).
Discussion
The distribution and prognostic significance of TP53 mutations found in this study based on Danish breast cancer patients is in accordance with previous publications Citation[1]. In addition, we found agreement between the recently detected distribution of the three TP53 codon 72 genotypes Arg/Arg, Pro/Pro and Arg/Pro and an earlier published distribution among Norwegian breast cancer patients Citation[15] and Danish controls Citation[27], the last indicating that codon 72 genotypes are not associated with risk of breast cancer development. However, the lack of association between the different codon 72 genotypes and the number of TP53 mutations found in this study is in contrast to the findings by Langerød et al. who demonstrated 28.5% of patients carrying Arg/Arg of codon 72 also carried a TP53 mutation but only 3.8% of patients carrying Pro/Pro Citation[15]. This skewed distribution was found in all their series of breast cancer patients, which included two consecutive series and two series of advanced breast cancer cases. The diverging findings could in theory be due to more advanced disease in the Norwegian patient cohort as compared to the Danish patient cohort. So, we stratified our data according to the Nottingham Prognostic Index and found a weak tendency in the poor prognosis group towards more patients with retention of Arg also carrying a TP53 mutation, however the tendency was not statistically significant. Neither did we recover any difference in disease-free survival among patients with different TP53 codon 72 genotypes. This is in discordance with the studies by Tommiska et al. Citation[13] and Goode et al. Citation[16]. However, these two studies reported completely opposite results, namely that the Pro allele was either associated with poor prognosis Citation[13] or with good prognosis Citation[16], which might indicate that the Arg72Pro genotype itself lacks prognostic relevance. Also stratifying for TP53 mutations did not recover any difference in survival. However, we cannot exclude that our study may include too few patients to identify a difference between the three genotypes. In addition, it must be stressed out that our patient cohort was a selected cohort from a larger consecutive cohort, which might leave us with a somehow skew distribution of the patients. In general most of the well-known prognostic clinical markers were significant in our selected cohort but remarkably estrogen receptor revealed no prognostic significance.
We are not aware of other studies investigating both, LOH of codon 72, specific allele retention, and TP53 mutation together in breast cancer. Bonafe et al. published a study regarding LOH of codon 72 and outcome Citation[17]. The study included 67 breast cancer patients with 29 patients heterozygous of codon 72. A significantly reduced survival was found for the 7 heterozygous patients with LOH and retention of Arg as compared to a combined group of heterozygous patients without LOH and homozygous patients. We did not confirm these findings. Actually, we found the opposite namely a poorer survival associated with patients with retention of the Pro allele. However this association was particularly evident among the 11 patients with TP53 mutations indicating that the prognostic significance of codon 72 LOH might depend on mutations in the TP53 gene. One might speculate that the discrepancy between the studies could result from our use of a non-conventional method estimating genotypes and LOH; however, running part of the samples with a PCR-RFLP based method confirmed our findings. Probably and more importantly, it must be kept in mind that both studies are based on a very limited number of patients and caution must be exercised interpreting the findings. In conclusion our findings suggest that the Arg72Pro polymorphism itself is neither significantly associated with TP53 mutations nor with disease-free survival. However, among heterozygous patients, LOH of codon 72 might be associated with TP53 mutations, and within the group of heterozygous patients retention of the Pro allele might be associated with a poor breast cancer prognosis as compared to retention of the Arg allele or no LOH. This association was, however, only statistically significant in the small group of 11 patients with TP53 mutations.
References
- Borresen-Dale AL. TP53 and breast cancer. Hum Mutat 2003; 21: 292–300
- Overgaard J, Yilmaz M, Guldberg P, Hansen LL, Alsner J. TP53 mutation is an independent prognostic marker for poor outcome in both node-negative and node-positive breast cancer. Acta Oncol 2000; 39: 327–33
- Alsner J, Yilmaz M, Guldberg P, Hansen LL, Overgaard J. Heterogeneity in the clinical phenotype of TP53 mutations in breast cancer patients. Clin Cancer Res 2000; 6: 3923–31
- Cho Y, Gorina S, Jeffrey PD, Pavletich NP. Crystal structure of a p53 tumor suppressor-DNA complex: Understanding tumorigenic mutations. Science 1994; 265: 346–55
- Walker KK, Levine AJ. Identification of a novel p53 functional domain that is necessary for efficient growth suppression. Proc Natl Acad Sci U S A 1996; 93: 15335–40
- Matlashewski GJ, Tuck S, Pim D, Lamb P, Schneider J, Crawford LV. Primary structure polymorphism at amino acid residue 72 of human p53. Mol Cell Biol 1987; 7: 961–3
- Papadakis EN, Dokianakis DN, Spandidos DA. p53 codon 72 polymorphism as a risk factor in the development of breast cancer. Mol Cell Biol Res Commun 2000; 3: 389–92
- Buyru N, Tigli H, Dalay N. P53 codon 72 polymorphism in breast cancer. Oncol Rep 2003; 10: 711–4
- Huang XE, Hamajima N, Katsuda N, Matsuo K, Hirose K, Mizutani M, et al. Association of p53 codon Arg72Pro and p73 G4C14-to-A4T14 at exon 2 genetic polymorphisms with the risk of Japanese breast cancer. Breast Cancer 2003; 10: 307–11
- Noma C, Miyoshi Y, Taguchi T, Tamaki Y, Noguchi S. Association of p53 genetic polymorphism (Arg72Pro) with estrogen receptor positive breast cancer risk in Japanese women. Cancer Lett 2004; 210: 197–203
- Mabrouk I, Baccouche S, El Abed R, Mokdad-Gargouri R, Mosbah A, Said S, et al. No evidence of correlation between p53 codon 72 polymorphism and risk of bladder or breast carcinoma in Tunisian patients. Ann N Y Acad Sci 2003; 1010: 764–70
- Helland A, Langerod A, Johnsen H, Olsen AO, Skovlund E, Borresen-Dale AL. p53 polymorphism and risk of cervical cancer. Nature 1998; 396: 530–1
- Tommiska J, Eerola H, Heinonen M, Salonen L, Kaare M, Tallila J, et al. Breast cancer patients with p53 Pro72 homozygous genotype have a poorer survival. Clin Cancer Res 2005; 11: 5098–103
- Han W, Kang D, Park IA, Kim SW, Bae JY, Chung KW, et al. Associations between breast cancer susceptibility gene polymorphisms and clinicopathological features. Clin Cancer Res 2004; 10: 124–30
- Langerod A, Bukholm IR, Bregard A, Lonning PE, Andersen TI, Rognum TO, et al. The TP53 codon 72 polymorphism may affect the function of TP53 mutations in breast carcinomas but not in colorectal carcinomas. Cancer Epidemiol Biomarkers Prev 2002; 11: 1684–8
- Goode EL, Dunning AM, Kuschel B, Healey CS, Day NE, Ponder BA, et al. Effect of germ-line genetic variation on breast cancer survival in a population-based study. Cancer Res 2002; 62: 3052–7
- Bonafe M, Ceccarelli C, Farabegoli F, Santini D, Taffurelli M, Barbi C, et al. Retention of the p53 codon 72 arginine allele is associated with a reduction of disease-free and overall survival in arginine/proline heterozygous breast cancer patients. Clin Cancer Res 2003; 9: 4860–4
- Sjogren S, Inganas M, Norberg T, Lindgren A, Nordgren H, Holmberg L, et al. The p53 gene in breast cancer: prognostic value of complementary DNA sequencing versus immunohistochemistry. J Natl Cancer Inst 1996; 88: 173–82
- Geisler S, Lonning PE, Aas T, Johnsen H, Fluge O, Haugen DF, et al. Influence of TP53 gene alterations and c-erbB-2 expression on the response to treatment with doxorubicin in locally advanced breast cancer. Cancer Res 2001; 61: 2505–12
- Xu Y, Yao L, Ouyang T, Li J, Wang T, Fan Z, et al. p53 Codon 72 polymorphism predicts the pathologic response to neoadjuvant chemotherapy in patients with breast cancer. Clin Cancer Res 2005; 11: 7328–33
- Offersen BV, Sorensen FB, Yilmaz M, Knoop A, Overgaard J. Chalkley estimates of angiogenesis in early breast cancer–relevance to prognosis. Acta Oncol 2002; 41: 695–703
- Hansen LL, Andersen J, Overgaard J, Kruse TA. Molecular genetic analysis of easily accessible breast tumour DNA, purified from tissue left over from hormone receptor measurement. APMIS 1998; 106: 371–7
- Hansen LL, Yilmaz M, Overgaard J, Andersen J, Kruse TA. Allelic loss of 16q23.2-24.2 is an independent marker of good prognosis in primary breast cancer. Cancer Res 1998; 58: 2166–9
- Wu G, Hua L, Zhu J, Mo QH, Xu XM. Rapid, accurate genotyping of beta-thalassaemia mutations using a novel multiplex primer extension/denaturing high-performance liquid chromatography assay. Br J Haematol 2003; 122: 311–6
- Baccouche S, Mabrouk I, Said S, Mosbah A, Jlidi R, Gargouri A. A more accurate detection of codon 72 polymorphism and LOH of the TP53 gene. Cancer Lett 2003; 189: 91–6
- Galea MH, Blamey RW, Elston CE, Ellis IO. The Nottingham Prognostic Index in primary breast cancer. Breast Cancer Res Treat 1992; 22: 207–19
- Hogdall EV, Hogdall CK, Christensen L, Glud E, Blaakaer J, Bock JE, et al. Distribution of p53 codon 72 polymorphisms in ovarian tumour patients and their prognostic significance in ovarian cancer patients. Anticancer Res 2002; 22: 1859–64