Abstract
Background: Lung cancer patients have a risk of recurrence even after curatively intended surgery. Cell-free circulating tumor DNA (ctDNA) and circulating tumor marker measurements are easily accessible through peripheral blood and could potentially identify patients with worse prognosis. The aim of this study was to examine ctDNA in pre-operative plasma and the role of tumor markers in pre-operative serum for their predictive potential on risk of tumor recurrence.
Methods: Mutation analysis by 26-gene targeted sequencing was performed on 157 lung adenocarcinomas (ACs) from patients surgically treated at the Lund University Hospital 2005–2014. Of these, 58 tumors from patients in stages I–IIIA (34 stage I, 14 stage II and 10 stage III) with mutation(s) in EGFR, BRAF or KRAS were included. ctDNA from corresponding plasma (median 1.5 ml, range 1–1.6) was analyzed for one tumor-specific mutation in either of these three oncogenes using ultrasensitive IBSAFE droplet digital PCR (ddPCR). The tumor markers cancer antigen 125 (CA 125) and carbohydrate antigen 19-9 (CA 19-9) were analyzed in corresponding serum with electrochemiluminiscence immunoassay.
Results: 6/7 patients with ctDNA and 19/51 without detected ctDNA were diagnosed with recurrence (log-rank test p = .001). 8/10 patients with positive serum tumor markers and 17/47 without tumor markers were diagnosed with recurrence (log-rank test, p = .0002). Fifteen patients had positive ctDNA and/or tumor markers, 12 of these had recurrence (log-rank test, p < .0001).
Conclusion: A combination of tumor markers and ctDNA single mutation detection in low-volume pre-operative blood samples is a promising prognostic test. Prediction of recurrent disease in surgically treated early stage lung cancer can likely be further improved by using larger volumes of blood.
Introduction
Non-small cell lung cancer (NSCLC) diagnosed at an early stage might be subjected to curatively intended lung cancer surgery. However, cancer recurrence occurs in 30–70% of operated NSCLC patients, with stage as an important prognostic factor [Citation1,Citation2]. Adjuvant chemotherapy has been shown to give an absolute increase in 5-year-survival by 4% in NSCLC [Citation3]. Easily accessible prognostic tests could make it possible to stratify tumors further into risk groups, aiming to individualize treatment and follow-up in order to improve lung cancer care. Blood-based tests could be a convenient, noninvasive way to reach that goal and a combination of different markers might add both sensitivity and specificity to such an approach.
Circulating protein tumor markers in serum (from here on referred to as tumor markers) may help in diagnosis and prognosis but a problem is lack of specificity and convincing results on how these markers are best used in relation to histology, stage and combinations of markers [Citation4]. In a previous study, we detected a trend of higher frequency of lung adenocarcinoma (AC) recurrence among patients with elevated CA 19-9 and CA 125 [Citation5].
Cell-free circulating tumor DNA (ctDNA) typically comprises a small fraction of the total circulating cell-free DNA (cfDNA) but even so it could potentially serve multiple clinical purposes [Citation6]. Analysis of ctDNA in plasma has not yet been implemented in the management of early stage lung cancer, but has already a complementary role in the care of patients with advanced disease, in order to detect tyrosine kinase inhibitor (TKI) sensitizing EGFR mutations or the resistance mutation T790M [Citation7]. Herein, we utilized plasma from patients with mutation positive tumors and performed ctDNA analysis with IBSAFE digital droplet PCR (ddPCR), an innovation upon standard ddPCR with enhanced sensitivity with a lower limit of detection of 0.001% variant allele frequency (VAF; George et al., manuscript in preparation).
In this retrospective study, we combined analysis of ctDNA in pre-operative plasma with analysis of tumor markers in pre-operative serum and compared the results with clinical outcome.
Patients and methods
Study inclusion
Patients with primary lung tumors of AC histology and with tumor samples (either formalin fixed paraffin embedded or fresh frozen) available for next-generation sequencing (NGS) were selected from the Southern Swedish Lung Cancer Study [Citation8] in which patients surgically treated for lung cancer at the Lund University Hospital were included (after written informed consent) between 2005 and 2014. None of the patients had neoadjuvant therapy. Tumor specimens, blood, serum and plasma have been collected and stored at −80 °C. Inclusion criteria for further blood-based analyses was mutation(s) in either of the genes EGFR, KRAS or BRAF.
Histopathology
Histology [Citation9], AC grade [Citation10], and pathological TNM according to the 7th edition [Citation11] were reviewed by a thoracic pathologist (HB). All tumors were originally classified as ACs. However, due to overlap with another study, one tumor has in parallel been re-classified as having a 10% large cell neuroendocrine tumor component and therefore classified as a combined large cell neuroendocrine cancer (LCNEC).
Adenocarcinoma grade 1 includes minimally invasive AC (non-mucinous/mucinous) and lepidic predominant AC, grade 2 includes acinary and papillary predominant AC and grade 3 includes micropapillary and solid predominant AC and invasive mucinous AC.
Clinical data
History of smoking, adjuvant treatment, date of lung cancer recurrence and death were collected from clinical files, ascertained through January 9 2019. Primary endpoint was recurrence-free interval (RFi) and events defined as recurrences were at date of radiological or pathological statement. Median follow-up time was calculated among recurrence-free patients as time from date of surgery to last/latest follow-up visit to the pulmonologist/oncologist. If a patient was diagnosed with metastatic spread from another primary cancer, the follow-up time was censored at time of diagnosis of the metastatic spread. Secondary endpoint was overall survival (OS).
Mutation analysis
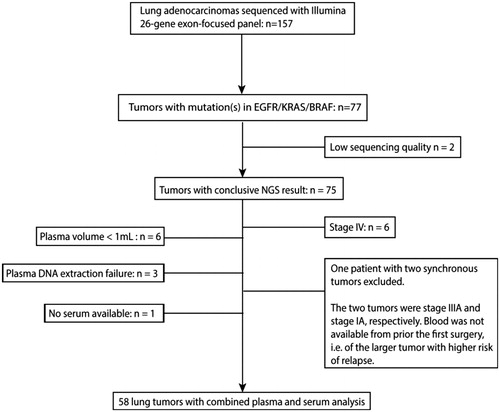
Specimens from 157 ACs from Southern Swedish Lung Cancer Study were analyzed with targeted NGS using the Illumina TruSightTumor 26-gene panel which is an exon-focused panel, performed as described previously [Citation12]. 77/157 tumor specimens harbored at least one mutation in either of the EGFR, BRAF or KRAS genes and 58 tumors were included in the study. Exclusion of samples are described in .
Figure 1. Study scheme, outlining inclusion of patients. Mutation analysis was performed on 157 surgically resected lung adenocarcinomas. Based on the tumors’ mutational status (KRAS-/EGFR-/BRAF-positive), stages (I–IIIA), and the availability of pre-operative blood samples, 58 patients were selected for analysis of plasma ctDNA and serum tumor markers.
Cell-free DNA extraction from plasma
cfDNA from 1000 to 1600 µl pre-operative plasma from the patients with mutation positive tumors were extracted using the Qiagen QIAamp Circulating Nucleic Acid kit, eluted in a total volume of 50 µl.
Plasma ctDNA analysis
IBSAFE ddPCR was used to analyze the plasma DNA for the specific mutation found in the corresponding tumor. IBSAFE is an improved method based upon ddPCR that enables improved lower limit of detection to 0.001% variant allele frequency assuming sufficient DNA copies are input (George et al., manuscript in preparation). Improved limit of detection is achieved by sequential combination of linear amplification to increase copies of true target sequence followed by limited exponential amplification for signal generation, thereby increasing sensitivity and specificity by drastically minimizing the consequence of polymerase base-incorporation errors. Custom IBSAFE assays targeting 23 unique mutations were designed based on the tumor NGS results. In this project, performance of the assays was verified using at least 500 ng of normal human DNA in negative control experiments, confirming a false-positive rate of 0 to 1 in 90,000 to 740,000 genome copies. At least two positive mutant droplets were considered a positive finding of ctDNA.
Tumor markers in serum
The tumor markers cancer antigen 125 (CA125) and carbohydrate antigen 19-9 (CA19-9) were analyzed in serum with electrochemiluminiscence immunoassay at the Division of Clinical Chemistry and Pharmacology, Department of Laboratory Medicine, Lund University, Lund, Sweden. Standard cutoff values for clinical testing (Skane University Hospital, Lund) were used: CA 19-9 < 35kE/L and CA 125 < 35kE/L.
Statistics
Statistical analysis was performed in R version 3.4.4 [Citation13]. The Kaplan–Meier method was used to compare RFi and OS between patient groups with different ctDNA and tumor marker status. Log-rank tests were used to calculate associations between recurrences and ctDNA and/or tumor markers. All tests were two-sided.
Results
Patients
Fifty-eight patients were included. Patient and tumor characteristics are shown in . Twenty-five of the patients were diagnosed with lung cancer relapse. 76% of recurrences were diagnosed within 3 years after surgery. Median follow-up for patients alive and without lung cancer relapse (n = 33) was 3.3 years (range 0.7–6.9). Three of the patients deceased before the final scheduled lung cancer-related follow-up. Their follow-up times were censored at time of the last follow-up visit. Furthermore, the follow-up time for one patient was censored at time of diagnosis of metastatic dissemination from a breast cancer initially treated with curative intention prior to lung cancer surgery (Supplementary Table 1).
Table 1. Baseline patient characteristics.
Two patients had mutations in two of the genes EGFR/KRAS/BRAF. Due to limits in blood sample volumes, and based on availability of assays developed previously in other projects, we chose only one mutation per case for further analysis in plasma with ddPCR. Mutations detected with NGS in tumors, mutation analyzed in plasma when two mutations were present, plasma volumes used for ddPCR analysis, and results from ddPCR and tumor marker analysis in serum are described together with detailed clinical data in Supplementary Table 1.
Tumor markers and ctDNA
Ten patients (17%) had pre-operative tumor markers in serum of which four had positive CA 19-9, four positive CA125 and two had a combination of positive CA 125 and CA 19-9. Of the 10 patients with tumor markers, eight (80%) were diagnosed with recurrence compared with 17/48 (35% of the patients with negative tumor markers (log rank test, p = .0002, ). Prevalence of tumor markers and recurrence in each stage separately is displayed in . Four of the patients with tumor markers had other malignancies in addition to their lung cancer (presented in Supplementary Table 1). One patient had bladder cancer with local recurrences and a prostate cancer without oncological treatment and another patient was diagnosed with breast cancer (T1N0M0) just over 1 year prior lung cancer. The third patient had bladder cancer with a local recurrence 2 years prior to lung cancer and the last one had skin tumors of squamous cell carcinoma type.
Figure 2. RFi and OS in relation to ctDNA and tumor marker status in pre-operative blood samples. (A) RFi in patients with and without tumor markers. (B) RFi in patients with detected ctDNA and no detected ctDNA. (C) RFi and (D) OS in patients with ctDNA and/or tumor markers or no detected ctDNA or tumor markers. Log-rank p-values are shown.
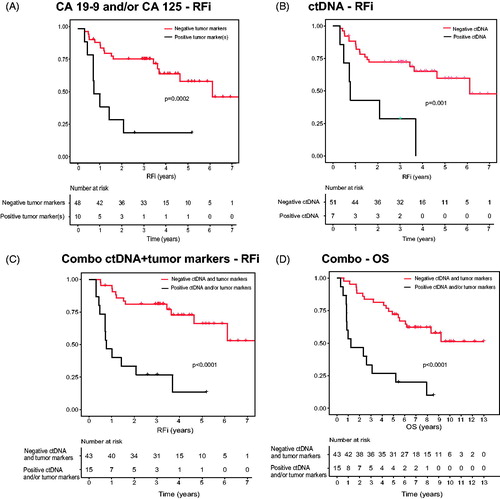
Table 2. Tumor markers and recurrence.
Analysis of ctDNA was performed on plasma samples with volumes between 1000 and 1600 µL. Total cfDNA concentration had a median 2078 copies/mL plasma (range 689–29391). Positive finding of mutant ctDNA with ddPCR was detected in 7/58 samples of pre-operative plasma, all with KRAS mutations and 4/7 with stage III tumors (). 6/7 patients with ctDNA in pre-operative plasma were diagnosed with lung cancer recurrence. Log rank test (p = .001) revealed a difference between ctDNA positive and negative patients (). There were too few results to allow for comparisons between stages.
Table 3. Samples with positive ctDNA in pre-operative plasma.
By combining single mutation detection in ctDNA and tumor marker analysis (positive CA 19-9 and/or CA 125) a positive result was demonstrated in 15 patients. Eight patients had positive CA 19-9 or CA 125 but no detected ctDNA, 5/15 had ctDNA but no tumor markers and the two patients with both tumor markers positive also had positive ctDNA. Twelve of these 15 patients positive for ctDNA and/or tumor markers had a lung cancer recurrence and there was a correlation between this combination approach and time to recurrence (log rank test, p < .0001; ). There was also a correlation between OS and the combination (log rank test, p < .0001; ).
Classification of AC grade revealed three tumors of grade 1, 39 tumors of grade 2 and 16 tumors of grade 3. Of stage I tumors (n = 34) 9% were grade 1, 68% grade 2 and 24% grade 3. Of stage II tumors (n = 14) none was grade 1, 86% were grade 2 and 14% grade 3 (including the tumor with re-classified histology to combined LCNEC). The smallest group, stage III (n = 10), consisted of no grade 1 tumors, 40% grade 2 and 60% grade 3. None of the three tumors of grade 1 (all stage I tumors) had tumor markers or positive ctDNA. Of the 39 tumors with grade 2, 4/39 (10%) had 1–2 tumor markers and 3/39 (7.7%) had positive ctDNA out of which one patient had both ctDNA and tumor markers. In tumors with grade 3, 6/16 (38%) had 1–2 tumor markers and 4/16 (25%) had positive ctDNA out of which one patient had both ctDNA, positive CA 125 and positive CA 19-9.
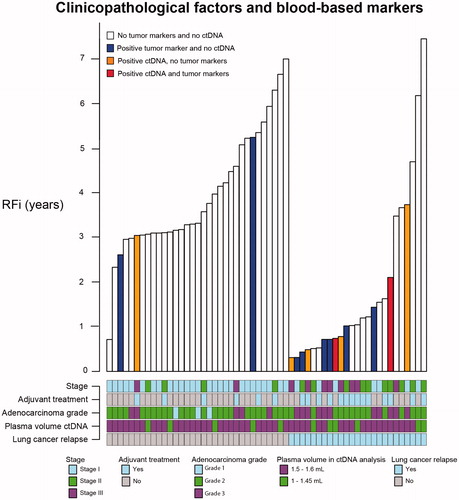
Stage, follow-up time, plasma volume, adjuvant treatment and AC grade are variables that might influence both ctDNA and tumor markers and the possible association with lung cancer relapse. These variables are illustrated in .
Figure 3. Clinicopathological factors and blood-based markers. Recurrence-free intervals (y-axis) are displayed in relation to results from ctDNA and tumor marker analyses (bar colours) in patients with subsequent lung cancer relapse (x-axis, to the right) or without lung cancer relapse (x-axis, left). Stage, plasma volume, adjuvant treatment and adenocarcinoma grade are described below each patient/bar.
Discussion
This study reveals the possible usefulness of blood-based biomarkers to identify surgically treated early-stage lung cancer patients with more aggressive tumors. In summary, we detected ctDNA in 7/58 (12%) pre-operative samples with a plasma volume range of 1.0–1.6 mL (median 1.5 mL) and by using a single mutation assay approach. Six of these seven patients with positive ctDNA in pre-operative plasma were later diagnosed with lung cancer recurrence. The one patient with positive ctDNA but no subsequent recurrence received adjuvant therapy and it is possible that the adjuvant therapy was of benefit for this patient. As previously described, adjuvant therapy in lung cancer has only a moderate overall impact on survival, with the weakest evidence in stage I disease, and biomarkers to identify the relatively few patients who benefit are lacking [Citation3]. Our results imply that larger studies on the potential use of pre-operative ctDNA for prediction of tumor aggressiveness and thereby benefit from adjuvant therapy are motivated. Patients with stage I disease would be of particular interest in such investigations, with the potential of changing today’s clinical decision-making. In the present study, with a limited population size, we noticed a tendency of clustering of tumor markers and ctDNA in higher stages, although not exclusively in higher stages (). This is consistent with results from a study of ctDNA across stages in different cancers other than lung cancer [Citation14]. Only 10 included patients had stage III disease, yet these patients constituted 4/7 cases with identified pre-operative ctDNA, suggesting that tumor stage is one important factor for identifying ctDNA. However, two of the positive ctDNA findings came from stage I patients (i.e. 2/34 stage I cases), and, interestingly, both patients were diagnosed with recurrence, after 0.7 and 3.7 years, respectively (). In a larger study population, it would be possible to analyze if tumor markers and/or ctDNA adds prognostic information to stage and thereby helps in decision-making in the adjuvant setting.
Importantly, a larger plasma volume may significantly improve ctDNA detection rates as the total number of input genome copy equivalents was relatively low, thereby the sample composition constrains the threshold on achievable limit of detection rather than the detection technology. A benefit of IBSAFE’s improved lower limit of detection is that potential background genomic DNA (e.g. from lysed leukocytes) has a very limited effect on the detection rate, although it may depress the measured VAFs. In the seven cases where we detected ctDNA in plasma, all harbored KRAS codon 12 mutations, thus in line with KRAS codon 12 mutation being the most common alteration within the 58 tumors as demonstrated by NGS. It requires further study whether this reflects mutation-specific biology. However, assay dependence is unlikely since all assays used for the IBSAFE ddPCR have been validated without demonstrating differences between mutations regarding sensitivity (George et al. manuscript in preparation).
Tracking multiple mutations instead of single driver mutation might affect the possibility of detecting ctDNA. Using multiplex-PCR NGS, Abbosh et al. [Citation15] tracked a median of 18 single nucleotide variants (SNV) in higher-volume plasma samples (median plasma volume 5 mL) with a threshold of two detected SNVs. In pre-operative plasma, ctDNA was detected in 46/96 early stage NSCLC and a single SNV in 12 additional cases. They also reported differences between histological subtypes. Detection rates were 11/58 (19%) in early stage lung ACs in total and 5/39 (13%) in stage I lung ACs. Detection rates in early stage squamous cell carcinomas (SqCC) were 30/31 (97%) in total and 16/17 (94%) in stage I. These results suggest that both stage and histology might influence ctDNA detection rates. We included only lung ACs, however, as stated previously, one of the ACs in our study was re-classified to combined LCNEC due to a 10% neuroendocrine component. Interestingly, this tumor had the highest amount of mutant ctDNA concentration and clearly higher than the other positive findings ().
Other variables associated with detection of ctDNA in the study by Abbosh et al. [Citation15] were necrosis, increased proliferative indices, lymphovascular invasion and tumor volume. We analyzed AC grade, which is another variable that might impact ctDNA concentration. As displayed in , positive ctDNA and tumor markers in our study cluster with both higher stage and higher grade. In contrast to Abbosh et al., Chen et al. [Citation16] did not reveal a difference in ctDNA detection across histologies. Chen et al. studied ctDNA and conventional tumor biomarkers in 76 stages I–IIIA NSCLC patients. In plasma from 10 mL peripheral blood (approximately 5 mL plasma equivalent) collected prior to surgery, they detected ctDNA with a 50-gene NGS panel in 48 samples (concordant with each mutation in tumor DNA in 31 samples, 10 samples with discordant mutations in plasma and tumor and 7 plasma samples with mutations not detected in the corresponding tumor). Of 59 ACs, 62.7% had positive ctDNA and 64.7% of 17 SqCC had positive ctDNA. Furthermore, the sensitivity of detection with ctDNA was 63.2% and sensitivity of detection with the tumor markers (CA 125, CA 19-9, CEA, neuron specific enolase and cytokeratin 19 fragment) was 49.3%.
Addition of tumor marker measurements in serum in our study identified more high-risk patients, i.e. patients that were later diagnosed with recurrence, than ctDNA alone in this study but also led to two more patients with positive values who were not diagnosed with lung cancer relapse. Protein tumor markers have been widely studied and, although not established in lung cancer management due to low sensitivity and specificity, they are sometimes used as a complement for e.g. diagnostic purposes [Citation17]. Here, we found a correlation between positive tumor markers and recurrence. A combination of pre-operative ctDNA and tumor markers detected more patients with eventual recurrence than ctDNA or tumor markers alone, although, due to a low number of positives and small amounts of plasma in our study, this needs to be further evaluated in larger cohorts.
This study has several limitations. It is a retrospective study with a limited number of patients. In general, metachronous lung cancer is not rare and can sometimes be difficult to distinguish from a recurrence. We have tried to minimize this risk by thorough review of radiology, histopathology, molecular pathology, and patients´ clinical charts. Another concern is plasma volume. We analyzed 1600 µL (1 case), 1500 µL (46 cases) and 1000–1450 µL (11 cases). With greater plasma volumes it is possible that we would have detected ctDNA in more samples, which could influence the correlation with recurrence. With more positive ctDNA findings and serial measurement during lung cancer follow-up it would also have been possible to relate concentration of mutated tumor DNA in plasma with clinical outcome. Furthermore, tumor markers in serum might be elevated due to other reasons than lung cancer, such as other malignancies or benign conditions. Optimal references might differ between lung cancer and other cancers.
In conclusion, a combination of blood-based biomarkers as predictors of recurrence deserves to be further investigated in larger cohorts with possibilities to evaluate accuracy of different combinations and the value in each stage separately. Even with limited plasma volumes we were able to detect ctDNA in early stage lung tumors, including stage I, with this sensitive single assay method IBSAFE. Our results therefore open up future prospective studies using this method to individualize post-surgical treatment and follow-up by detecting mutations in ctDNA in combination with tumor markers in serum. Furthermore, a panel of blood-based markers complementing radiology and clinical examination at post-operative follow-up could potentially lead to earlier detection of recurrence. Impact on blood-based markers of different variables such as histology, grade, and other tumor related or clinical factors should be further studied.
Supplemental Material
Download MS Excel (28.1 KB)Acknowledgments
All patients for their participation in the Southern Lung Cancer Study. All personnel at ward 1, Skane University Hospital in Lund, and at Department of Clinical Pathology in Lund, for their work with study inclusion and sample collection. Lena Mårtensson and Carolina Andersson Rios for administrative help.
Disclosure statement
AMG and LHS are named inventors of patent applications covering IBSAFE and are co-founders of SAGA Diagnostics AB. The other authors declare no conflicts of interest.
Additional information
Funding
References
- Cruz C, Afonso M, Oliveiros B, et al. Recurrence and risk factors for relapse in patients with non-small cell lung cancer treated by surgery with curative intent. Oncology. 2017;92:347–352.
- Consonni D, Pierobon M, Gail MH, et al. Lung cancer prognosis before and after recurrence in a population-based setting. J Natl Cancer Instit. 2015;107:djv059.
- Arriagada R, Auperin A, Burdett S, et al. Adjuvant chemotherapy, with or without postoperative radiotherapy, in operable non-small-cell lung cancer: two meta-analyses of individual patient data. Lancet (London, England). 2010;10375:1267–1277.
- Nakamura H, Nishimura T. History, molecular features, and clinical importance of conventional serum biomarkers in lung cancer. Surg Today. 2017;47:1037–1059.
- Isaksson S, Jonsson P, Monsef N, et al. CA 19-9 and CA 125 as potential predictors of disease recurrence in resectable lung adenocarcinoma. PloS One. 2017;12:e0186284.
- Santarpia M, Liguori A, D’Aveni A, et al. Liquid biopsy for lung cancer early detection. J Thorac Dis. 2018;10:S882–s897.
- Lindeman NI, Cagle PT, Aisner DL, et al. Updated molecular testing guideline for the selection of lung cancer patients for treatment with targeted tyrosine kinase inhibitors: Guideline From the College of American Pathologists, the International Association for the Study of Lung Cancer, and the Association for Molecular Pathology. Arch Pathol Lab Med. 2018;142:321–346.
- Brunnström H, Johansson L, Jirström K, et al. Immunohistochemistry in the differential diagnostics of primary lung cancer: an investigation within the Southern Swedish Lung Cancer Study. Am J Clin Pathol. 2013;140:37–46.
- Travis WD BE, Burke AP, Marx A, Nicholson AG, eds. WHO Classification of tumours of the lung, pleura, thymus and heart. 4th ed. Lyon, France: IARC Press; 2015.
- Travis WD, Brambilla E, Noguchi M, et al. International association for the study of lung cancer/American thoracic society/European respiratory society international multidisciplinary classification of lung adenocarcinoma. J Thoracic Oncol. 2011;6:244–285.
- Sobin L GM WC, eds. International Union Against Cancer (UICC). TNM classification of malignant tumours. 7th ed. Chichester, UK: Wiley-Blackwell; 2009.
- Lindquist KE, Karlsson A, Leveen P, et al. Clinical framework for next generation sequencing based analysis of treatment predictive mutations and multiplexed gene fusion detection in non-small cell lung cancer. Oncotarget. 2017;238:34796–34810.
- RC Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. Available from: https://www.R-project.org/2018.
- Bettegowda C, Sausen M, Leary RJ, et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med. 2014;6:224ra24.
- Abbosh C, Birkbak NJ, Wilson GA, et al. Phylogenetic ctDNA analysis depicts early-stage lung cancer evolution. Nature. 2017;545:446–451.
- Chen K, Zhang J, Guan T, et al. Comparison of plasma to tissue DNA mutations in surgical patients with non-small cell lung cancer. J Thoracic Cardiovasc Surg. 2017;154:1123–1131.e2.
- Zhao H, Chen KZ, Hui BG, et al. Role of circulating tumor DNA in the management of early-stage lung cancer. Thorac Cancer. 2018;9:509–515.
