Abstract
Background: Remodelling and spacing factor 1 (RSF1) is involved in the regulation of chromatin remodelling and represents a potential therapeutic target. High RSF1 expression has been linked to adverse tumour features in many cancer types, but its role in prostate cancer is uncertain.
Methods: In this study, RSF1 expression was analysed by immunohistochemistry on a tissue microarray with 17,747 prostate cancers.
Results: Nuclear RSF1 staining of 16,456 interpetable cancers was considered strong, moderate, weak and negative in 25.2%, 48.7%, 5.3% and 20.8% of cancers respectively. Positive RSF1 expression was associated with advanced tumour stage, high Gleason grade, lymph node metastasis (p < .0001 each), early biochemical recurrence (p < .0003) and more frequent in the ERG positive than in the ERG negative subset (88% versus 71%; p < .0001). Subset analysis revealed, that associations between RSF1 expression and unfavourable tumour phenotype and PSA recurrence were present in both subgroups but stronger in the ERG negative than in the ERG positive subset. The univariate Cox proportional hazard ratio for PSA recurrence-free survival for strong versus negative RSF1 expression was a weak 1.60 compared with 5.91 for the biopsy Gleason grade ≥4 + 4 versus ≤3 + 3. The positive association of RSF1 protein detection with deletion of 3p13, 10q23 (PTEN), 12p13, 16q23, and 17p13 (p < .0001 each) suggest a role of high RSF1 expression in the development of genomic instability.
Conclusion: In summary, the results of our study identify RSF1 as an independent prognostic marker in prostate cancer with a particularly strong role in ERG negative cases.
Introduction
Prostate cancer is common in elder men in Western societies [Citation1]. Most of them grow slowly but a small subset can be highly aggressive requiring intensified treatment [Citation2,Citation3]. Predictive prognostic parameters include Gleason grade and tumour extent on biopsies, serum prostate-specific antigen (PSA) levels, and clinical stage. While powerful in retrospective analysis of large cohorts, they are suboptimal for individual treatment decisions. Therefore it is hoped that identification of clinically applicable molecular markers will improve prediction of prostate cancer aggressiveness.
Remodelling and spacing factor 1 (RSF1) alias hepatitis B virus X protein associated protein is of potential clinical interest [Citation4]. RSF1 is involved in the regulation of chromatin remodelling. It is a ubiquitously expressed histone chaperone, that interacts with specific chromatin remodelling factors [Citation5]. Chromatin-remodelling complexes play a key role in all processes that require a change of the chromatin structure including DNA repair, DNA synthesis, and mitosis. RSF1 was reported to become overexpressed in various cancers including ovarian [Citation6], lung [Citation7], hepatocellular [Citation8] or colon cancer [Citation9]. Studies in ovarian [Citation6], lung [Citation7], nasopharyngeal [Citation10], gastric [Citation11], rectal [Citation12] and urinary bladder cancer [Citation13] suggested that its overexpression might be linked to poor patient prognosis. RSF1 overexpression was also found to potentially contribute to paclitaxel resistance [Citation14], and cancers recurring after chemotherapy or radiotherapy showed higher RSF1 values than their respective primary tumours [Citation15]. A role of RSF1 in modulating drug sensitivity was also supported by experiments showing that RSF1 knockdown by siRNA treatment reduced cell growth, increased drug sensitivity, and induced cell death in cancer cells with RSF1 overexpression [Citation14,Citation16].
Based on the analysis of 169 patients, a link between RSF1 overexpression and high tumour stage and poor prognosis was recently also suggested for prostate cancer [Citation17]. To better understand the clinical and biological impact of RSF1 expression in prostate cancer, an immunohistochemical analysis of RSF1 expression was performed on a cohort of more than 17,000 prostate cancer specimens in a tissue microarray (TMA) format.
Material and methods
Patients
The 17,747 patients had radical prostatectomy between 1992 and 2014 at the Department of Urology and the Martini Clinic at the University Medical Centre Hamburg-Eppendorf. The prostate specimen was completely embedded for histological analysis [Citation18]. Classical Gleason categories and “quantitative” Gleason grading was performed as described before [Citation19]. Follow-up data were available for a total of 14,464 patients (median 48 months, range: 1 to 276 months; ). Prostate specific antigen (PSA) recurrence was defined as the time point when the postoperative PSA level was at least 0.2 ng/ml and increasing at subsequent measurements. The TMA was produced as described and contained various control tissues, including normal prostate tissue [Citation20]. It was annotated with data on ERG expression, and ERG break apart FISH analysis [Citation21] and deletion status of 3p13 (FOXP1) [Citation22]), 5q21 (CHD1) [Citation23], 6q15 (MAP3K7) [Citation24], 8p21 (NKX3) [Citation25], 10q23 (PTEN) [Citation26]), 12p13 (CDKN1B) [Citation27], 12q24 (NCOR2) [Citation28], 16q23 (WWOX) [Citation27], 17p13 (TP53) [Citation29], 18q21 (NEDD4L) [Citation30], and Ki67 labelling index (Ki67-LI) data [Citation31].
Table 1. Characteristics of the arrayed prostate cancers.
Immunohistochemistry
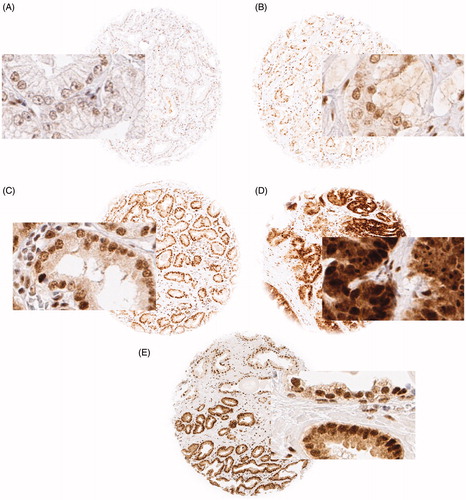
Freshly cut TMA sections were stained in one experiment. Slides were dewaxed and exposed to 121 °C for 5 min in pH 7.8 Tris-EDTA buffer. Anti-RSF1 antibody [EPR3749(2)] (rabbit monoclonal antibody ab109002, Abcam, Great Britain) was applied at 1:4050 and 37 °C for 60 min. Bound antibody was visualised (EnVision Kit, Dako, Glostrup, Denmark) according to the manufacturer’s directions. RSF1 staining of variable intensity was predominantly seen in the nucleus and was occasionally accompanied by a weaker cytoplasmatic co-staining. For this study, the nuclear staining intensity was estimated as negative, weak, moderate or strong for each tumour containing TMA spot ().
Figure 1. Representative pictures of (A) negative, (B) weak, (C) moderate and (D) strong RSF1 nuclear staining in prostate cancer and (E) in a mixed spot with normal glands in the upper half and cancerous glands in the lower half; magnification 100×, spot size 600 μm.
Statistical analysis
Contingency tables were calculated to study association between RSF1 expression and clinico-pathological variables, and the chi-square test was used to find significant relationships. Ki67 labelling data were tested by ANOVA. Kaplan-Meier curves were generated using PSA recurrence as the clinical endpoint. The log-rank test was applied to test the significance of differences between stratified survival functions. Cox proportional hazards were calculated for biochemical relapse in univariate and multivariate models to compare various predictive parameters and tested by chi-square test. JMP 12 (SAS Institute Inc., NC, USA) was used. P-values are uncorrected for multiple testing.
Results
The TMA analysis was interpretable in 16,456 of 17,747 tumour samples (93%). Non-informative cases (7%) lacked tissue samples or unequivocal cancer tissue in the TMA spot. Normal prostate epithelial tissue showed medium to strong nuclear RSF1 staining. In cancers, RSF1 staining was localised mostly in the nucleus and/or in cytoplasm. Detectable nuclear RSF1 staining was seen in 13,037 of our 16,456 (79.2%) interpretable tumours and was considered weak in 5.3%, moderate in 54.0% and strong in 25.2% of cancer. The remaining 3419 (20.8%) tumours were negative for RSF1. Representative images of RSF1 staining are given in .
Associations with ERG-status and tumour phenotype
The intensity and the presence of nuclear RSF1 staining were increased in ERG positive or ERG rearranged tumours (Figure S1) and showed associations with advanced tumour stage (p < .0001), and high Gleason grade (p < .0001 ). The latter associations held true in the subset of ERG negative cancers (Table S1) and ERG positive cancers (Table S2).
Table 2. Association between RSF1 staining and prostate cancer phenotype.
Association to other key genomic deletions
Comparison of RSF1 expression with deletions of 3p13, 5q21 (CHD1), 6q15, 8p21, 10q23 (PTEN), 12p13, 12q24, 16q23, 17p13, and 18q21 revealed that RSF1 staining was significantly positive linked to all of these deletions but 5q21, 6q15 and 18q21 (Figure S2). The strongest positive association was observed for 10q23 (PTEN). When Bonferroni-corrected for multiple comparisons, p-values remained significant except for 12q24.
Association to tumour cell proliferation (Ki67-LI)
RSF1 staining intensity was significantly associated with increased cell proliferation as measured by Ki67-LI (Table S3). These associations were independent from ERG fusion status (p < .0001) and the Gleason grade, as they also held true in subgroups of tumours with identical Gleason score (≤3 + 3, 3 + 4, 4 + 3; p < .0001 each and ≥4 + 4; p = .0112 each).
Association with PSA recurrence
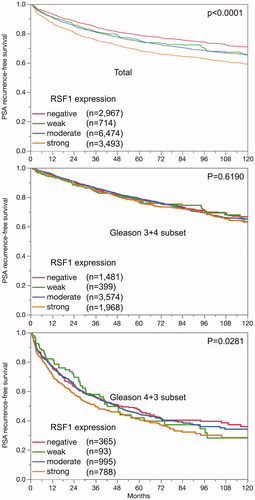
Follow-up data were available from 13,465 patients with interpretable RSF1 staining. The intensity of nuclear RSF1 staining was associated with early PSA recurrence after prostatectomy (p < .0001, ). The PSA relapse rate of 25% was observed after 87, 61, 58 and 37 month in patients with negative, weak, moderate and strong RSF1 expression. The findings were independent of the ERG status (data not shown). When stratified for Gleason group 2 (3 + 4) and 3 (4 + 3) RSF1 expression had no impact on clinical outcome in the former but in the latter ().
Uni- and multivariate analysis of hazard ratio
The clinical relevance of nuclear RSF1 expression was evaluated in 4 scenarios (Table S4). Nuclear RSF1 expression provided highly significant prognostic value beyond the established pre- and postoperative parameters in all scenarios for all cancers and the ERG negative subset. There was a weaker independent prognostic role for RSF1 in the ERG positive subset for the preoperative scenario 4 (). In uni- and multivariate analysis the prognostic effect of RSF1 compared with other markers was weak (hazard ratio of 1.60 respective 1.43).
Table 3. Cox proportional hazards for PSA recurrence-free survival after prostatectomy of established preoperative prognostic parameter and RSF1 expression.
Discussion
The results of our study demonstrate that increased nuclear RSF1 expression is a weak but independent predictor of poor prognosis, especially in the ERG negative subset of prostate cancer.
Detectable levels of RSF1 expression were found in 79.2% of 16,456 interpretable prostate cancers. This is comparable to the findings of Li et al. who described overexpression of RSF1 in 45% of 169 prostate cancers [Citation17]. RSF1 protein may be involved in cancer progression, as its expression levels further increased with advanced pathological stage, higher Gleason score, lymph node metastasis, elevated tumour cell proliferation and early PSA recurrence. Li et al. had also suggested a possible link of RSF1 overexpression with unfavourable prostate cancer phenotype. A tumour promoting role of RSF1 overexpression is further supported by several other studies describing a relationship between RSF1 overexpression and poor patient outcome in ovarian [Citation6], rectal [Citation12], urinary bladder [Citation13], lung [Citation7], and breast cancer [Citation32] as well as be various experimental models [Citation14–16].
The availability of molecular data from earlier studies utilising this prostate carcinoma TMA enabled us to investigate the relationship of RSF1 expression with molecularly defined cancer subgroups, the most relevant of which are ERG positive and ERG negative cancers. TMPRSS2:ERG fusions occur in about 50% of prostate cancers and lead to a constitutive overexpression of the transcription factor ERG. ERG overexpression by itself lacks prognostic relevance, but ERG modulates the expression of more than 1600 genes in prostate epithelial cells [Citation21,Citation33,Citation34]. The prognostic effect of RSF1 expression was slightly stronger in ERG negative than in ERG positive cancers but was also retained in the latter group (Table S1). A modified cellular microenvironment may play a causative role for the reduced prognostic role of RSF1 expression in ERG positive cancers. Our assay sensitivity may serve as an alternative explanation for different prognostic effects between ERG positive and ERG negative cancers. The diagnostic window where differences in the expression level can be assessed by brightfield IHC is relatively narrow. All tumours below a certain threshold are diagnosed as “negative”, all tumours above a certain expression level are considered strongly positive. It is thus possible, that our immunohistochemistry protocol was better suited to distinguish expression differences in cancers with generally lower expression levels (ERG negative cancers) than in those with higher expression (ERG positive cancers). Irrespective of its cause, the different prognostic impact of RSF1 in ERG positive and negative cancers demonstrates, that the applicability (and perhaps thresholds) of prognostic markers may depend on specific molecular tumour features. This represents a considerable challenge for the development of prognostic cancer tests that shall be applicable to every patient.
Chromosomal deletions account for the next ten most common recurrent genomic alterations in prostate cancer after TMPRSS2:ERG fusions. These deletions may define clinically relevant molecular subtypes of prostate cancer. Most of them are either linked to ERG positive (10q (PTEN), 3p13, 8p21, 17p13 (TP53), 12q24) or ERG negative cancer (6q15, 5q21 (CHD1), 16q23) [Citation28,Citation35,Citation36]. That most of these deletions were related to RSF1 expression in ERG positive and ERG negative tumours may indicate an involvement of RSF1 in controlling genomic integrity or double strand breakage repair [Citation37,Citation38]. The known role of RSF1 as a chromatin remodelling protein is consistent with RSF1 overexpression impacting genomic instability. This is in line with previous studies showing that RSF1 overexpression induces the endogenous DNA damage response by activating the ATM signalling pathway [Citation38]. RSF1 overexpression increased the levels of the endogenous DNA damage signalling pathway and impaired efficient repair upon DNA damage [Citation37].
RSF1 analysis provided limited additional prognostic information beyond the established preoperative and postoperative prognostic parameters in prostate cancer. It is important, however, that prognostic parameters are needed for prostate cancer patients that are not only independent of established factors, but also better reproducible to be suitable for prospective analysis. Most routinely used prognostic features suffer from significant limitations. The quality of the lymph node status data greatly depends on the extent of surgery and the pathological work-up of the removed materials [Citation39]. The Gleason Grade, the most powerful preoperatively available prognostic marker, suffers from substantial interobserver variability reaching beyond 40% in individual biopsies [Citation40]. In the present retrospective TMA study two pathologists analysed a single spot from each tumour and discrepant cases were decided by a third one. Whether this can be implemented in the clinical routine for prospective testing of multiple representative samples for a patient to deal with the heterogeneity of prostate cancer remains to be seen.
Conclusion
The results of the study identify nuclear RSF1 staining as a weak but independent prognosticator in prostate cancer.
Ethics approval and consent to participate
The ethics committee of the Ärztekammer Hamburg approved this study (WF-049/09). According to local laws (HmbKHG, §12a) informed consent was not required for this study.
Supplemental Material
Download MS Word (5.6 MB)Acknowledgment
The authors appreciate the excellent technical support of Christina Koop, Janett Lütgens, Sünje Seekamp, and Inge Brandt.
Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability
All data generated or analysed during this study are included in this published article [and its supplementary information files].
References
- Torre LA, Bray F, Siegel RL, et al. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65(2):87–108.
- Wilt TJ, Brawer MK, Jones KM, et al. Radical prostatectomy versus observation for localized prostate cancer. N Engl J Med. 2012;367(3):203–213.
- Thompson IM, Jr, Tangen CM. Prostate cancer–uncertainty and a way forward. N Engl J Med. 2012;367(3):270–271.
- Lee TH, Elledge SJ, Butel JS. Hepatitis B virus X protein interacts with a probable cellular DNA repair protein. J Virol. 1995;69(2):1107–1114.
- Shamay M, Barak O, Shaul Y. HBXAP, a novel PHD-finger protein, possesses transcription repression activity. Genomics. 2002;79(4):523–529.
- Shih I-M, Sheu JJ-C, Santillan A, et al. Amplification of a chromatin remodeling gene, Rsf-1/HBXAP, in ovarian carcinoma. Proc Natl Acad Sci U S A. 2005;102(39):14004–14009.
- Zhang X, Fu L, Xue D, et al. Overexpression of Rsf-1 correlates with poor survival and promotes invasion in non-small cell lung cancer. Virchows Arch. 2017;470(5):553–560.
- Xie C, Fu L, Xie L, et al. Rsf-1 overexpression serves as a prognostic marker in human hepatocellular carcinoma. Tumor Biol. 2014;35(8):7595–7601.
- Liu S, Dong Q, Wang E. Rsf-1 overexpression correlates with poor prognosis and cell proliferation in colon cancer. Tumor Biol. 2012;33(5):1485–1491.
- Tai HC, Huang HY, Lee SW, et al. Associations of Rsf-1 overexpression with poor therapeutic response and worse survival in patients with nasopharyngeal carcinoma. J Clin Pathol. 2012;65(3):248–253.
- Hu BS, Yu HF, Zhao G, et al. High RSF-1 expression correlates with poor prognosis in patients with gastric adenocarcinoma. Int J Clin Exp Pathol. 2012;5(7):668–673.
- Lin CY, Tian YF, Wu LC, et al. Rsf-1 expression in rectal cancer: with special emphasis on the independent prognostic value after neoadjuvant chemoradiation. J Clin Pathol. 2012;65(8):687–692.
- Liang PI, Wu LC, Sheu JJ, et al. Rsf-1/HBXAP overexpression is independent of gene amplification and is associated with poor outcome in patients with urinary bladder urothelial carcinoma. J Clin Pathol. 2012;65(9):802–807.
- Choi JH, Sheu JJ, Guan B, et al. Functional analysis of 11q13.5 amplicon identifies Rsf-1 (HBXAP) as a gene involved in paclitaxel resistance in ovarian cancer. Cancer Res. 2009;69(4):1407–1415.
- Fang FM, Li CF, Huang HY, et al. Overexpression of a chromatin remodeling factor, RSF-1/HBXAP, correlates with aggressive oral squamous cell carcinoma. Am J Pathol. 2011;178(5):2407–2415.
- Sheu JJ, Choi JH, Yildiz I, et al. The roles of human sucrose nonfermenting protein 2 homologue in the tumor-promoting functions of Rsf-1. Cancer Res. 2008;68(11):4050–4057.
- Li H, Zhang Y, Zhang Y, et al. Rsf-1 overexpression in human prostate cancer, implication as a prognostic marker. Tumor Biol. 2014;35(6):5771–5776.
- Schlomm T, Iwers L, Kirstein P, et al. Clinical significance of p53 alterations in surgically treated prostate cancers. Mod Pathol. 2008;21(11):1371–1378.
- Sauter G, Steurer S, Clauditz TS, et al. Clinical utility of quantitative gleason grading in prostate biopsies and prostatectomy specimens. Eur Urol. 2016;69(4):592–598.
- Kononen J, Bubendorf L, Kallionimeni A, et al. Tissue microarrays for high-throughput molecular profiling of tumor specimens. Nat Med. 1998;4(7):844–847.
- Minner S, Enodien M, Sirma H, et al. ERG status is unrelated to PSA recurrence in radically operated prostate cancer in the absence of antihormonal therapy. Clin Cancer Res. 2011;17(18):5878–5888.
- Krohn A, Seidel A, Burkhardt L, et al. Recurrent deletion of 3p13 targets multiple tumour suppressor genes and defines a distinct subgroup of aggressive ERG fusion-positive prostate cancers. J Pathol. 2013;231(1):130–141.
- Burkhardt L, Fuchs S, Krohn A, et al. CHD1 is a 5q21 tumor suppressor required for ERG rearrangement in prostate cancer. Cancer Res. 2013;73(9):2795–2805.
- Kluth M, Hesse J, Heinl A, et al. Genomic deletion of MAP3K7 at 6q12-22 is associated with early PSA recurrence in prostate cancer and absence of TMPRSS2:ERG fusions. Mod Pathol. 2013;26:975–983.
- Kluth M, Amschler NN, Galal R, et al. Deletion of 8p is an independent prognostic parameter in prostate cancer. Oncotarget. 2017;8:379–392.
- Krohn A, Diedler T, Burkhardt L, et al. Genomic deletion of PTEN is associated with tumor progression and early PSA recurrence in ERG fusion-positive and fusion-negative prostate cancer. Am J Pathol. 2012;181(2):401–412.
- Kluth M, Runte F, Barow P, et al. Concurrent deletion of 16q23 and PTEN is an independent prognostic feature in prostate cancer. Int J Cancer. 2015;137(10):2354–2363.
- Weischenfeldt J, Simon R, Feuerbach L, et al. Integrative genomic analyses reveal an androgen-driven somatic alteration landscape in early-onset prostate cancer. Cancer Cell. 2013;23(2):159–170.
- Kluth M, Harasimowicz S, Burkhardt L, et al. Clinical significance of different types of p53 gene alteration in surgically treated prostate cancer. Int J Cancer. 2014;135(6):1369–1380.
- Kluth M, Graunke M, Moller-Koop C, et al. Deletion of 18q is a strong and independent prognostic feature in prostate cancer. Oncotarget. 2016;7(52):86339–86349.
- Minner S, Jessen B, Stiedenroth L, et al. Low level HER2 overexpression is associated with rapid tumor cell proliferation and poor prognosis in prostate cancer. Clin Cancer Res. 2010;16(5):1553–1560.
- Aydin OZ, Vermeulen W, Lans H. ISWI chromatin remodeling complexes in the DNA damage response. Cell Cycle. 2014;13(19):3016–3025.
- Brase JC, Johannes M, Mannsperger H, et al. TMPRSS2-ERG -specific transcriptional modulation is associated with prostate cancer biomarkers and TGF-beta signaling. BMC Cancer. 2011;11(1):507.
- Tomlins SA, Rhodes DR, Perner S, et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310(5748):644–648.
- Taylor BS, Schultz N, Hieronymus H, et al. Integrative genomic profiling of human prostate cancer. Cancer Cell. 2010;18(1):11–22.
- Lapointe J, Li C, Giacomini CP, et al. Genomic profiling reveals alternative genetic pathways of prostate tumorigenesis. Cancer Res. 2007;67(18):8504–8510.
- Min S, Choi YW, Yun H, et al. Post-translational regulation of the RSF1 chromatin remodeler under DNA damage. Mol Cells. 2018;41(2):127–133.
- Sheu JJ, Guan B, Choi JH, et al. Rsf-1, a chromatin remodeling protein, induces DNA damage and promotes genomic instability. J Biol Chem. 2010;285(49):38260–38269.
- Wilczak W, Wittmer C, Clauditz T, et al. Marked prognostic impact of minimal lymphatic tumor spread in prostate cancer. Eur Urol. 2018;74(3):376–386.
- Egevad L, Ahmad AS, Algaba F, et al. Standardization of Gleason grading among 337 European pathologists. Histopathology. 2013;62(2):247–256.