Abstract
Background: Statins treat hyperlipidemia and prevent cardiovascular morbidity and mortality. Evidence suggests that they also have anti-neoplastic activity. Several studies show a reduced rate of breast cancer recurrence among lipophilic statin users (e.g., simvastatin), motivating calls for clinical trials of statins in breast cancer patients. We measured the impact of genetic variation in statin-metabolizing enzymes and drug transporters on the recurrence rate in simvastatin-treated breast cancer patients.
Methods: We conducted a nested case-control study among Danish women diagnosed with non-metastatic, invasive breast cancer between 2004–2010 who had filled ≥1 prescription for simvastatin after diagnosis. Cases were all breast cancer recurrences from the source population; one control was matched to each case on cancer stage, estrogen receptor and hormone therapy status, calendar period of diagnosis, and duration of simvastatin exposure. We genotyped variants in simvastatin-metabolizing enzymes (CYP3A4/rs35599367 and CYP3A5/rs776746) and drug transporters (ABCB1/rs2032582 and SLCO1B1/rs4149056), and estimated their association with recurrence with logistic regression models.
Results: We observed protective (though imprecisely-measured) associations between variants in genes encoding drug transporters (ABCB1 and SLCO1B1) and simvastatin-metabolizing enzymes (CYP3A4 and CYP3A5) and breast cancer recurrence in simvastatin-treated women. For example, carrying two variant alleles in ABCB1 was associated with a 31% lower rate of recurrence (multivariable OR = 0.69, 95% CI: 0.31, 1.5).
Conclusion: Our study provides weak evidence to support the use of genetic variation in ABCB1, SLCO1B1, CYP3A4, and CYP3A5 as biomarkers of breast tumor response to simvastatin. Validation of these findings within adjuvant clinical trials is encouraged.
Introduction
Statins (HMG-CoA reductase inhibitors) are prescribed to reduce levels of low density lipoprotein cholesterol (LDL-C) and to thereby prevent cardiovascular morbidity and mortality. Accumulating evidence suggests these drugs also have adjuvant anti-cancer effects [Citation1]. Statins are orally administered, inexpensive, well-tolerated, and chemically stable under most storage conditions. If proven effective as cancer therapeutic agents, statins could profoundly improve cancer survival worldwide. Statins have been well studied in relation to breast cancer incidence and survival. While most epidemiologic studies show no association between statin use and breast cancer incidence [Citation2–4], there is strong non-randomized evidence for a protective effect of statins on breast cancer recurrence [Citation5–7]. This evidence motivated calls for a clinical trial of statins in the breast cancer adjuvant setting [Citation6,Citation8,Citation9], and such trials are currently underway (for example, clinicaltrials.gov numbers NCT03971019, NCT00816244).
Previous studies have shown that the protective association between statins and breast cancer recurrence is not modified by important breast tumor prognostic characteristics, such as estrogen receptor expression, stage, and histological grade [Citation10]. Nonetheless, host and tumor molecular features may influence individual response to statin treatment. Among these are the genes encoding proteins that metabolize and transport statins, some of which harbor functional polymorphisms that affect statin disposition and impact either the effectiveness of statins for cholesterol lowering or the incidence of adverse drug reactions [Citation11–13]. We hypothesized that these same functional polymorphisms would modify the protective association between simvastatin (the most commonly prescribed statin in our study’s Danish source population [Citation10]) and breast cancer recurrence. Such a result would suggest a predictive marker of adjuvant anti-cancer effectiveness, and would also bolster the evidence base supporting statins' adjuvant potential.
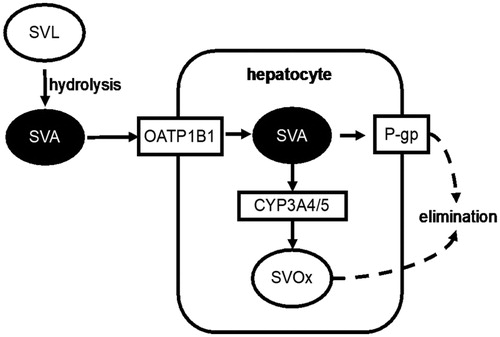
illustrates the disposition of simvastatin. Ingested simvastatin lactone (SVL) is hydrolyzed in the gut to simvastatin acid (SVA), the active form of the drug. SVA absorbed into the bloodstream enters hepatocytes via the organic anion transporter 1B1 (OATP1B1), where it interrupts the mevalonate pathway and inhibits the production of LDL-C. Within hepatocytes, SVA is either metabolized to inactive oxidized products (SVOx) by the CYP3A4 and CYP3A5 enzymes (CYP3A4/5), or excreted into the bile by the P-glycoprotein transporter (P-gp). OATP1B1 is encoded by the SLCO1B1 gene, which harbors a single-nucleotide polymorphism that reduces enzyme function (rs4149056) and consequently increases the plasma concentrations of transporter substrates. We hypothesized that the protective association between simvastatin exposure and breast cancer recurrence would be stronger in carriers of this variant allele. SNPs in CYP3A4 and CYP3A5 (rs35599367 and rs776746, respectively) impair or eliminate expression of the CYP3A4/5 enzymes, resulting in reduced clearance of substrate drugs. We hypothesized that carriers of any of the CYP3A4/5 variants would have a stronger response to simvastatin with respect to breast cancer recurrence prevention. The ABCB1 gene, which encodes P-gp, has a triallelic SNP (rs2032582) with variant alleles causing attenuated gene expression, leading to a slower rate of biliary excretion of SVA. The resulting buildup of SVA inside hepatocytes eventually generates a concentration gradient sufficient to drive passive diffusion of SVA into the plasma [Citation14]. We hypothesized that carriers of any rs2032582 variant would have greater protection against recurrence from simvastatin exposure. We evaluated these hypotheses by conducting a Danish population-based case-control study of breast cancer patients who filled prescriptions for simvastatin while at risk for recurrence.
Figure 1. Transporter proteins and metabolic enzymes involved in the disposition of simvastatin. Administered simvastatin lactone (SVL) is hydrolyzed in the gut to simvastatin acid (SVA). SVA enters hepatocytes via OATP1B1 (encoded by the polymorphic SLCO1B1 gene). SVA is oxidized by CYP3A4 and CYP3A5 (encoded by eponymous polymorphic genes) into oxidized metabolites (SVOx), which are eliminated. In addition, the P-glycoprotein transporter (P-gp; encoded by the polymorphic ABCB1 gene) drives efflux of SVA from hepatocytes into the bile.
Methods
Source population
The source population included all female residents of Denmark diagnosed with non-metastatic, invasive breast cancer between 2004–2010 and who filled at least one prescription for simvastatin after diagnosis (n = 846).
Data sources and enumeration of cases and controls
Our study combined data from several Danish population-based medical and sociodemographic registries [Citation15]. These registries can be unambiguously linked at the individual level using the civil registration system number (CPR), a unique personal identifier assigned to all legal residents of Denmark upon birth or immigration [Citation16,Citation17]. Cases were identified from the Danish Breast Cancer Group (DBCG) Registry [Citation18], and defined as women from the source population who were diagnosed with a local, regional, or distant breast cancer recurrence during the study period. Given available study resources and the cost of genotyping, we targeted an enrollment of approximately 150 recurrence cases from this source population. For each enrolled case we enumerated a risk set comprised of all women from the source population who were alive and recurrence-free after the same amount of follow-up time and who matched the case on breast cancer stage (I, II, or III), estrogen receptor (ER) and endocrine therapy (ET) status, calendar period of initial breast cancer diagnosis (2004–2006, 2007–2008, and 2009–2010), and duration of simvastatin exposure between diagnosis and index date (±25%). We matched on duration of simvastatin exposure during the recurrence risk period to ensure that genetic associations would be estimable within conditionally uniform backdrops of statin treatment. Data on vital status came from the Danish Civil Registration System [Citation16], and data on simvastatin prescriptions came from the Danish National Prescription Registry [Citation19]. We selected one control at random from each case’s risk set. Index dates of cases (the date of recurrence diagnosis) were assigned to their matched controls.
Tissue procurement, DNA extraction, and genotyping
We queried the Danish Pathology Data Bank [Citation20] to identify hospitals at which cases and controls had their primary breast tumors resected. Pathology departments from these hospitals then retrieved archived formalin-fixed, paraffin-embedded (FFPE) breast tumor blocks and shipped them to the Institute of Pathology at Aarhus University Hospital for DNA extraction. Tissue blocks were handled according to a strict standard operating procedure to prevent cross-contamination and nuclease digestion. Three to six sections (10 μm thickness) were removed from each tumor block and transferred to sterile 1.5 mL polypropylene microcentrifuge tubes. Paraffin was removed with xylene, after which DNA was extracted with two incubations in 99% ethanol. Contaminating proteins were digested with proteinase K, and DNA was purified using the QIAamp DNA FFPE Tissue Kit (Qiagen AB, Dusseldorf, Germany) according to the manufacturer’s protocol.
Commercially-available TaqMan kits (Applied Biosystems, Thermo Fisher Scientific, Foster City, California, USA) were used to assay variants in ABCB1 (rs2032582; assay IDs: C_11711720C_30 and C_11711720D_40), CYP3A4 (rs35599367; assay ID: C_59013445_10), CYP3A5 (rs776746; assay ID: C_26201809_30) and SLCO1B1 (rs4149056; assay ID: C_30633906_10). All assays were performed on a ViiA7 real-time PCR machine (Applied Biosystems, Thermo Fisher Scientific, Foster City, California, USA). For TaqMan assays, 30 ng of purified DNA were amplified in 10 µL PCR reactions. PCR reactions were incubated at 60 °C for 30 s and 95 °C for 10 min, then cycled 60 times between 15 s incubations at 95 °C and 60 s incubations at 60 °C. Genotypes were classified based on TaqMan VIC/FAM intensity values using the autocall feature of the QuantStudio Software V1.3. For the CYP3A5 (rs776746) variant, genotypes of selected samples (including all samples genotyped as heterozygous) were confirmed using Sanger sequencing.
Definitions of analytic variables
We defined breast cancer recurrence as diagnosis with a local, regional, or distant recurrence, or contralateral breast cancer, per DBCG convention. Duration of simvastatin exposure was defined as the number of days between breast cancer diagnosis and the end of follow-up that were covered by all filled simvastatin prescriptions, calculated based on fill quantity, tablet strength, and the defined daily dose (DDD) for simvastatin (30 mg) [Citation21]. ER status was based on pathology reports from treating hospitals and ascertained from the DBCG registry. Adjuvant endocrine therapy for ER-positive patients was recommended based on the DBCG treatment protocol in place at the time of a patient’s diagnosis. Our study period was covered by two treatment protocols, both of which specified 5 years of tamoxifen treatment for premenopausal women. Postmenopausal women diagnosed before January 207 would also have been recommended to 5 years of tamoxifen. Thereafter, postmenopausal women (including those previously started on 5 years of tamoxifen) were recommended to 5 years of sequential therapy comprised of 2 to 3 years of tamoxifen followed by an aromatase inhibitor [Citation22,Citation23]. Duration of follow-up was the number of days elapsed between incident diagnosis and the index date. We defined the following set of candidate confounders: age at the time of initial breast cancer diagnosis was categorized for descriptive purposes, but modeled as a continuous variable; menopausal status at initial breast cancer diagnosis was defined as pre- or post-menopausal as recorded in the DBCG registry; Union for International Cancer Control (UICC) summary stage was categorized as I, II, or III and modeled as a factor variable to account for matching of controls to cases; type of breast cancer surgery was classified as either breast-conserving or mastectomy; tumor ER status and receipt of adjuvant endocrine therapy (ET) were factored into a single variable with categories of ER-negative tumor and no endocrine therapy (ER−/ET−, the reference category), ER-positive tumor and no endocrine therapy (ER+/ET−), and ER-positive tumor with endocrine therapy (ER+/ET+), modeled as a factor variable to account for matching of controls to cases; chemotherapy was dichotomized into received or did not receive; and duration of simvastatin exposure during the recurrence risk period was categorized for description as ≤1 year, 2–3 years, 4–5 years, and ≥6 years, but treated as a continuous variable to account for matching of controls to cases. Genotypes for ABCB1, CYP3A4, CYP3A5, and SLCO1B1 were initially defined as the number of variant alleles (0, 1, or 2) carried by a patient, with homozygous wild-type (0 variant alleles) serving as the reference category. The rarity of homozygous variant genotypes in CYP3A4, CYP3A5, and SLCO1B1 led us to collapse genotypes into “≥1 variant allele” versus “0 variant alleles”, and to combine the related CYP3A4 and CYP3A5 genes into a single variable, CYP3A4/5, in which the comparison was between “≥1 variant allele” (i.e., any number of variants in rs35599367 and rs776746) and the reference category of “0 variant alleles.”
Statistical analysis
We calculated expected genotype frequencies under Hardy-Weinberg equilibrium and compared these with observed frequencies using Haldane’s exact test [Citation24]. We tabulated the frequency and proportion of breast cancer recurrence cases and matched controls according to demographic and clinical characteristics. We fit both conditional and multivariable logistic regression models to estimate associations between candidate biomarker genotypes and recurrence in simvastatin-treated breast cancer patients. Conditional models accounted for the matching factors (UICC stage, ER/ET status, calendar period of initial diagnosis, and duration of simvastatin exposure), while multivariable models included matching factors and additionally adjusted for age, receipt of chemotherapy, and type of breast cancer surgery. Multivariable modeling allowed us to break matched pairs and analyze the entire set of cases and controls who were successfully genotyped. As a consequence of the risk-set sampling of controls, the odds ratios (OR) from these models estimate the incidence rate ratio from the hypothetical underlying cohort [Citation25].
Results
Genotyping performance
shows the observed and expected minor allele frequencies and genotype categories among our study’s controls. Observed minor allele frequencies were very similar to published benchmarks for Caucasian populations in the dbSNP database (https://www.ncbi.nlm.nih.gov/snp/), and observed genotype frequencies were very similar to those predicted under Hardy-Weinberg equilibrium. Together, these indicate genotyping of high technical quality.
Table 1. Summary of gene variant characteristics and their distribution among controls.
Characteristics of cases and controls
We enrolled 151 recurrence cases among simvastatin-exposed breast cancer patients. One control was selected for each of these cases, but 4 of these had insufficient tissue available for DNA extraction and were later excluded. Analyses were therefore based on 151 cases and 147 controls. reports the frequency and proportion of cases and controls according to key demographic and clinical characteristics. Compared with controls, recurrence cases were somewhat older () and were less likely to have received adjuvant chemotherapy (21% of cases vs. 27% of controls). Cases and controls were similar with respect to menopausal status, stage of the incident breast cancer, type of surgery received, ER/ET status, and duration of simvastatin exposure while at risk for recurrence ().
Table 2. Characteristics of breast cancer recurrence cases and matched controls. Denmark, 2004–2010.
Breast cancer recurrence associations for candidate biomarkers of simvastatin response
reports the conditional and multivariable-adjusted associations between functional polymorphisms in ABCB1, CYP3A4/5, and SLCO1B1 and recurrence in simvastatin-exposed breast cancer patients. All odds ratio point estimates were below unity—indicating a lower rate of recurrence among carriers of variant alleles in these genes compared with carriers of normal alleles—but associations were imprecisely measured, as seen by the widths of the 95% confidence intervals. Carrying two variant alleles in ABCB1 (rs2032582) was associated with a 31% lower rate of breast cancer recurrence (multivariable OR = 0.69, 95% CI: 0.31, 1.5); carrying at least one variant allele in CYP3A4/5 was associated with a 28% lower rate of breast cancer recurrence (multivariable OR = 0.72, 95% CI: 0.38, 1.4); and carrying at least one variant allele in SLCO1B1 was associated with a 20% lower rate of recurrence (multivariable OR = 0.80, 95% CI: 0.45, 1.4). Associations were similar between conditional and multivariable models, except in the case of CYP3A5, where the recurrence association for the rs776745 variant was substantially attenuated in the multivariable model.
Table 3. Associations between variants in simvastatin transport and metabolism genes and breast cancer recurrence among simvastatin-exposed breast cancer patients. Denmark, 2004–2010.
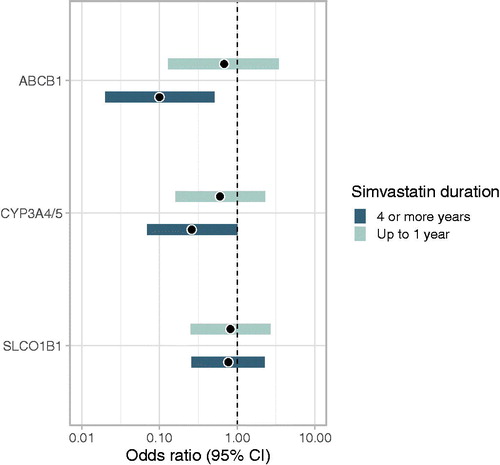
shows associations between gene variants and recurrence within strata representing the shortest (up to 1 year) and longest (4 or more years) durations of simvastatin exposure while at risk for recurrence. All associations moved further in the protective direction in the longer duration stratum, compared with the shorter duration stratum, which lends further credence to the hypothesis that these variants modify an effect of simvastatin on breast cancer recurrence.
Figure 2. Associations between variants in simvastatin transport and metabolism genes and breast cancer recurrence among simvastatin-exposed breast cancer patients, stratified by duration of simvastatin use after breast cancer diagnosis. Denmark, 2004–2010.
ABCB1: ORs compare ‘2 variant alleles’ group with ‘no variant alleles’ group.
CYP3A4/5 and SLCO1B1: ORs compare ‘≥1 variant allele’ group with ‘no variant alleles’ group.
Discussion
We observed lower rates of breast cancer recurrence among simvastatin-exposed breast cancer patients who carried variant alleles in genes for transporters (ABCB1 and SLCO1B1) and metabolic enzymes (CYP3A4 and CYP3A5) involved in the disposition of simvastatin. Though associations were imprecisely measured, our findings are consistent with established evidence linking these same gene variants with both cholesterol lowering response to simvastatin treatment and the incidence of outcomes (some adverse) related to higher extrahepatic statin activity. The rs2032582 variant alleles in ABCB1 are associated with greater LDL cholesterol reduction in simvastatin-treated patients, consistent with the expected effect of reducing the rate of SVA elimination [Citation13]. The rs35599367 and rs776746 variants in CYP3A4 and CYP3A5 are associated with lower required statin doses for achieving clinical lipid control benchmarks, which is also consistent with a reduced rate of SVA elimination. The SLCO1B1 rs4149056 variant is associated with higher relative area under the curve and Cmax for SVA [Citation26], and also with the risk of statin-induced myopathy, an adverse drug reaction that depends on extrahepatic action of statins [Citation27]. Higher extrahepatic activity and decreased rates of SVA elimination may also potentiate action against breast tumors, as suggested by the lower recurrence rates of variant carriers in our candidate genes.
The chief limitation of our study is the low precision with which recurrence associations were measured. The modest sample size also precluded fitting models in which genetic variant associations were mutually adjusted, so we could not explore how multiple variants interact to modify recurrence rate. We also prioritized genotyping the major, clinically important variants in the candidate genes, and could not define protein function phenotypes based on comprehensive genotype information. It is possible that some patients classified as carrying two normal alleles in fact carried one or more copies of an unmeasured variant allele. If the functional impact of the unmeasured variant(s) was to reduce protein function, then our estimates of association would be biased toward the null. It is also possible that relying on tumor tissue as the DNA source may have led to misclassification of germline genotype due to somatic mutations specific to the tumor and/or due to loss of heterozygosity at the SNP loci [Citation28–30]. Germline genotype is the important biological concept in our study, as the mechanisms involved in simvastatin disposition operate in the liver, not in the tumor tissue. The excellent quality of our genotyping data—both in terms of observed and expected allele frequencies and the lack of excess homozygotes according to the Hardy-Weinberg analysis—argue against these phenomena as sources of bias in our study. We note that the duration of simvastatin exposure among cases and controls varied substantially, with approximately half of subjects having a year or less of exposure during their recurrence risk period (). This may have reduced the magnitude of our observed associations, compared with an ideal scenario in which initiation would uniformly occur soon after diagnosis. Finally, due to resource limitations we did not enroll a series of recurrence cases and controls who were not exposed to simvastatin in order to rule out direct effects of the variant alleles on breast cancer recurrence. However, we are not aware of any published evidence linking these variants with recurrence risk, nor are there compelling biological hypotheses that would lead us to suspect such a link.
The major strengths of our study are its unselected Danish source population (which virtually eliminates the possibility of selection bias) [Citation31] and its use of high-quality registry data on simvastatin exposure [Citation19,Citation32], vital status [Citation16,Citation17], and breast tumor characteristics, treatment, and recurrence [Citation33,Citation34]. We expect little to no residual confounding, as germline genetic status is encoded at conception, and thus cannot be acted upon causally by demographic, behavioral, or clinical factors that may influence recurrence risk [Citation35]. This expectation is supported by the lack of change in associations upon adjustment for very strong recurrence risk factors (e.g., receipt of chemotherapy), which were not associated with metabolic enzyme and transporter genotypes.
In summary, our study weakly supports the hypothesis that functional polymorphisms in ABCB1, CYP3A4/5, and SLCO1B1 potentiate the putative anti-recurrence effect of statins—specifically simvastatin—in breast cancer patients. We recommend further study of these and other functional variants in the same genes to improve understanding of patient response. We further recommend that new studies evaluate non-genetic candidate modifiers of the statin/recurrence association, including breast tumor expression of HMG-CoA-reductase and the low-density lipoprotein receptor [Citation36].
Disclosure statement
None reported.
Additional information
Funding
References
- Chan KK, Oza AM, Siu LL. The statins as anticancer agents. Clin Cancer Res. 2003;9(1):10–19.
- Bonovas S, Filioussi K, Tsavaris N, et al. Use of statins and breast cancer: a meta-analysis of seven randomized clinical trials and nine observational studies. J Clin Oncol. 2005;23(34):8606–8612.
- Undela K, Srikanth V, Bansal D. Statin use and risk of breast cancer: a meta-analysis of observational studies. Breast Cancer Res Treat. 2012;135(1):261–269.
- Borgquist S, Tamimi RM, Chen WY, et al. Statin use and breast cancer risk in the nurses’ health study. Cancer Epidemiol Biomarkers Prev. 2016;25(1):201–206.
- Manthravadi S, Shrestha A, Madhusudhana S. Impact of statin use on cancer recurrence and mortality in breast cancer: a systematic review and meta-analysis. Int J Cancer. 2016;139(6):1281–1288.
- Ahern TP, Lash TL, Damkier P, et al. Statins and breast cancer prognosis: evidence and opportunities. Lancet Oncol. 2014;15(10):e461–8.
- Borgquist S, Bjarnadottir O, Kimbung S, et al. Statins: a role in breast cancer therapy? J Intern Med. 2018;284(4):346–357.
- Kumar A, Campbell M, Benz C, et al. A call for clinical trials: lipophilic statins may prove effective in treatment and prevention of particular breast cancer subtypes. J Clin Oncol. 2006;24:2127.
- Holmes MD, Chen WY. Hiding in plain view: the potential for commonly used drugs to reduce breast cancer mortality. Breast Cancer Res. 2012;14(2):216.
- Ahern TP, Pedersen L, Tarp M, et al. Statin prescriptions and breast cancer recurrence risk: a Danish nationwide prospective cohort study. J Natl Cancer Inst. 2011;103(19):1461–1468.
- Kitzmiller JP, Sullivan DM, Phelps MA, et al. CYP3A4/5 combined genotype analysis for predicting statin dose requirement for optimal lipid control. Drug Metab Drug Interact. 2013;28:59–63.
- Sadee W. Gene-gene-environment interactions between drugs, transporters, receptors, and metabolizing enzymes: statins, SLCO1B1, and CYP3A4 as an example. J Pharm Sci. 2013;102(9):2924–2929.
- Fiegenbaum M, da Silveira FR, Van der Sand CR, et al. The role of common variants of ABCB1, CYP3A4, and CYP3A5 genes in lipid-lowering efficacy and safety of simvastatin treatment. Clin Pharmacol Ther. 2005;78(5):551–558.
- Sharma P, Butters CJ, Smith V, et al. Prediction of the in vivo OATP1B1-mediated drug-drug interaction potential of an investigational drug against a range of statins. Eur J Pharm Sci. 2012;47(1):244–255.
- Schmidt M, Schmidt SAJ, Adelborg K, et al. The Danish health care system and epidemiological research: from health care contacts to database records. Clin Epidemiol. 2019;11:563–591.
- Pedersen C, Gøtzsche H, Møller J, et al. The Danish Civil Registration System. A cohort of eight million persons. Dan Med Bull. 2006;53(4):441–449.
- Schmidt M, Pedersen L, Sorensen HT. The Danish Civil Registration System as a tool in epidemiology. Eur J Epidemiol. 2014;29(8):541–549.
- Blichert-Toft M, Christiansen P, Mouridsen HT. Danish Breast Cancer Cooperative Group - DBCG: history, organization, and status of scientific achievements at 30-year anniversary. Acta Oncol. 2008;47(4):497–505.
- Pottegård A, Schmidt SAJ, Wallach-Kildemoes H, et al. Data resource profile: the Danish national prescription registry. Int J Epidemiol. 2017;46(3):798–798.
- Erichsen R, Lash T, Hamilton-Dutoit S, et al. Existing data sources for clinical epidemiology: the Danish National Pathology Registry and Data Bank. Clin Epidemiol. 2010;2:51–56.
- Guidelines for ATC Classification and DDD Assignment. Oslo, Norway: WHO Collaborating Centre for Drug Statistics Methodology; 2019 [accessed 2019 July 12]. Available from: https://www.whocc.no/filearchive/publications/2019_guidelines_web.pdf.
- Moller S, Jensen MB, Ejlertsen B, et al. The clinical database and the treatment guidelines of the Danish Breast Cancer Cooperative Group (DBCG); its 30-years experience and future promise. Acta Oncol. 2008;47(4):506–524.
- Jensen M-B, Laenkholm A-V, Offersen BV, et al. The clinical database and implementation of treatment guidelines by the Danish Breast Cancer Cooperative Group in 2007–2016. Acta Oncol. 2018;57(1):13–18.
- Schaid DJ, Batzler AJ, Jenkins GD, et al. Exact tests of Hardy-Weinberg equilibrium and homogeneity of disequilibrium across strata. Am J Hum Genet. 2006;79(6):1071–1080.
- Rothman KJ, Greenland S., Lash TL Case-control studies. In: Rothman KJ, Greenland S, Lash TL, editors. Modern epidemiology. 3rd ed. Philadelphia (PA): Lippincott, Williams & Wilkins; 2008. p. 111–127.
- Choi HY, Bae K-S, Cho S-H, et al. Impact of CYP2D6, CYP3A5, CYP2C19, CYP2A6, SLCO1B1, ABCB1, and ABCG2 gene polymorphisms on the pharmacokinetics of simvastatin and simvastatin acid. Pharmacogenet Genomics. 2015;25(12):595–608.
- Wilke RA, Ramsey LB, Johnson SG, et al. The clinical pharmacogenomics implementation consortium: CPIC guideline for SLCO1B1 and simvastatin-induced myopathy. Clin Pharmacol Ther. 2012;92(1):112–117.
- Xie B, Freudenheim J, Cummings S, et al. Accurate genotyping from paraffin-embedded normal tissue adjacent to breast cancer. Carcinogenesis. 2006;27(2):307–310.
- Osborne RJ, Hamshere MG. A genome-wide map showing common regions of loss of heterozygosity/allelic imbalance in breast cancer. Cancer Res. 2000;60(14):3706–3712.
- Miller B, Wang D, Krahe R, et al. Pooled analysis of loss of heterozygosity in breast cancer: a genome scan provides comparative evidence for multiple tumor suppressors and identifies novel candidate regions. Am J Hum Genet. 2003;73(4):748–767.
- Frank L. EPIDEMIOLOGY: when an entire country is a cohort. Science. 2000;287(5462):2398–2399.
- Riis AH, Erichsen R, Ostenfeld EB, et al. Validating registry data on statins prescriptions by blood measurements. Pharmacoepidemiol Drug Saf. 2019;28(5):609–615.
- Collin LJ, Cronin-Fenton DP, Ahern TP, et al. Cohort profile: the Predictors of Breast Cancer Recurrence (ProBe CaRE) premenopausal breast cancer cohort study in Denmark. BMJ Open. 2018;8(7):e021805.
- Cronin-Fenton DP, Kjaersgaard A, Ahern TP, et al. Validity of Danish Breast Cancer Group (DBCG) registry data used in the predictors of breast cancer recurrence (ProBeCaRe) premenopausal breast cancer cohort study. Acta Oncol. 2017;56(9):1155–1160.
- Rothman KJ, Greenland S, Lash T. Validity in epidemiologic studies. In: Rothman KJ, Greenland S, Lash TL, editors. Modern epidemiology. 3rd ed. Philadelphia (PA): Wolters Kluwer; 2008. p. 128–147.
- Gelissen IC, McLachlan AJ. The pharmacogenomics of statins. Pharmacol Res. 2014;88:99–106.
