Abstract
Background
To determine the impact of programed death-ligand 1 (PD-L1) expression on progression-free survival (PFS) outcomes in stage IV epidermal growth factor receptor (EGFR)-mutated non-small cell lung cancer (NSCLC) treated with first-line EGFR tyrosine kinase inhibitors (TKIs).
Material and methods
We searched biomedical databases for studies comparing PFS outcomes of PD-L1-positive versus (vs) PD-L1-negative tumors. We assessed the methodological quality of eligible studies using ROBINS-I tool. We employed a two-staged meta-analysis approach by reconstructing individual patient data of each study from the published Kaplan-Meier curves and then pooling the individual hazard ratios (HRs) and weighted mean differences (WMDs) for restricted mean PFS time at 6 (RMPFST6) and 12 (RMPFST12) months using random-effect models. We assessed the quality of summarized evidence using GRADE approach.
Results
We identified five non-randomized comparative studies including 435 patients. The overall risk of bias in the methodological quality of included studies was moderate. PD-L1-positive tumors were associated with significantly worse PFS outcomes compared to PD-L1-negative tumors (HR: 2.41, 95% confidence interval (CI): 1.59–3.66, p < .001; WMD in RMPFST6: −1.01, 95% CI: −1.65 to −0.37, p = .002; WMD in RMPFST12: −2.64, 95% CI: −4.40 to −0.88, p = .003). Subgroup analysis showed that the effect of PD-L1 expression on PFS outcomes was greater for studies using older-generation rather than third-generation TKIs (HR: 2.69 vs 1.22, p = .069; WMD in RMPFST6: −1.23 vs −0.07, p = .005; WMD in RMPFST12: −3.29 vs −0.12, p = .003). The quality of summarized evidence was judged to be low.
Conclusion
There is low certainty in the evidence to suggest that positive PD-L1 expression is associated with inferior disease control and survival outcomes in patients with stage IV EGFR-mutated NSCLC treated with first-line EGFR TKIs.
Introduction
The discovery of targeted therapy and immune checkpoint inhibitors has revolutionized the treatment landscape of advanced non-small cell lung cancer (NSCLC). The current standard first-line systemic therapy for newly-diagnosed advanced NSCLC harboring epidermal growth factor receptor (EGFR) mutation is EGFR tyrosine kinase inhibitors (TKIs) [Citation1].
Multiple randomized controlled trials (RCT) have shown that PD-L1 is a predictive biomarker for treatment response in patients with advanced NSCLC treated with immune checkpoint inhibitors [Citation1–4]. For example, KEYNOTE-042 demonstrated a survival benefit with the use of first-line pembrolizumab monotherapy over chemotherapy in patients with advanced NSCLC with a PD-L1 tumor proportion score (TPS) of 50% or greater [Citation1,Citation2].
Studies evaluating the frequencies of PD‐L1 expression in NSCLC harboring driver oncogenes have shown that EGFR mutation and PD‐L1 expression might be mutually present at the same time. The reported prevalence of positive PD-L1 expression in EGFR-mutated tumors ranges from 32% to 51% and 5 to 10% at the PD-L1 TPS of at least 1% and 50% respectively [Citation5,Citation6]. Investigators hypothesized that this population may represent a subset of EGFR mutants where the disease biology is more aggressive with the aberration of multiple oncogenic pathways and associated with various regulatory mechanisms causing TKI resistance [Citation7,Citation8]. However, the clinical evidence demonstrating the negative impact of PD-L1 expression on the survival outcomes of patients with EGFR-mutated NSCLC treated with first-line EGFR TKI is inconclusive. Previous clinical studies have reported conflicting findings [Citation5,Citation9,Citation10], which may be due to differences in prior systemic treatments, PD-L1 scoring cutoffs used and the types of antibody clones used [Citation11].
Hence, we aimed to perform a systematic review and meta-analysis to investigate the impact of PD-L1 expression on the survival outcomes of patients with stage IV EGFR-mutated NSCLC treated with first-line EGFR TKIs.
Material and methods
Study eligibility criteria
We included randomized or non-randomized comparative studies comparing the survival outcomes of PD-L1-positive and PD-L1-negative tumors in treatment-naïve patients diagnosed with histologically- or cytologically-proven stage IV EGFR-mutated NSCLC. Progression-free survival (PFS) was defined as time from EGFR TKI initiation to disease progression or death from any cause. Overall survival (OS) was defined as time from EGFR TKI initiation to death from any cause. EGFR status was confirmed by polymerase chain reaction method on either tissue or blood. PD-L1 status was confirmed by immunohistochemistry (IHC) method. Studies that used IHC method to determine EGFR status or defined survival outcomes from time of diagnosis or evaluated EGFR TKIs as second or subsequent lines of treatment were excluded.
Search strategy
We searched MEDLINE via PUBMED and EMBASE from January 2015 to February 2020 for relevant studies. The search strategy included the medical subject headings of ‘Carcinoma, Non-small cell lung cancer’, ‘Lung neoplasms’, ‘ErbB Receptors’, ‘Receptor, Epidermal Growth Factor’, ‘Antigens, CD274’. ‘Programmed cell death 1 receptor’ and ‘protein kinase inhibitors’ (Supplementary Data Citation1). The results were then hand searched for eligible studies. In addition, the reference lists of the selected studies were examined for any other relevant studies. We followed PRISMA reporting guideline [Citation12].
Study selection and data extraction
We used COVIDENCE, an online software, for study screening and selection [Citation13]. Two reviewers independently assessed the eligibility of abstracts identified by the search. The full-text papers of the studies that appeared to meet the inclusion criteria were retrieved and scrutinized. Disagreements were resolved by consensus.
The same reviewers extracted the data independently using standardized data collection forms. Data collected from the studies included publication details, study design, methodologic components and study characteristics, such as sample size, median age, outcome measures (PFS and OS, if reported), type of EGFR TKI used, PD-L1 positivity threshold and type of antibody clones used.
Methodologic quality assessment
We adopted Risk Of Bias In Non-Randomized Studies of Interventions (ROBINS-I) tool to assess the methodological quality of individual studies [Citation14]. ROBINS-I tool is used to examine the risk of bias of non-randomized studies based on the following domains: bias due to confounding, bias in selection of participants into the study, bias in classifications of interventions, bias due to deviations from intended interventions, bias due to missing data, bias in measurement of the outcome and bias in selection of reported result [Citation15]. The judgments within each domain carry forward to an overall risk of bias judgment across all domains. The overall risk of bias judgment can be categorized as low, moderate, serious and critical risk of bias. A low risk of bias suggests that the study is comparable to a well-performed randomized trial. A moderate risk of bias suggests that the study provides sound evidence for a non-randomized study but cannot be considered comparable to a well performed randomized trial. A serious risk of bias suggests that the study has some important problems while a critical risk of bias suggests that the study is too problematic to provide any useful evidence and should not be included in any synthesis. We planned to use the Cochrane Risk of Bias tool 2.0 (RoB) to assess the methodologic quality of randomized trials, but our search did not find any eligible randomized trials [Citation16].
Quality of evidence assessment
The quality of the overall evidence was assessed using the Grading of Recommendations, Assessment, Development and Evaluation (GRADE) approach [Citation15,Citation17,Citation18]. The domains assessed include study design, study’s overall risk of bias, inconsistency and imprecision of studies’ results and indirectness of the evidence. Other considerations include presence of publication bias, magnitude of effect and dose-response relationship. The GRADE system classifies the overall quality of the summarized evidence in one of the four grades: high, moderate, low and very low. A high-grade score suggests that further research is very unlikely to change our confidence in the estimate of effect and a very low-grade score indicates that any estimate of effect is very uncertain.
Outcomes of interest
The outcomes of interest in this study were PFS and OS. PFS was defined as time from EGFR TKI treatment initiation to disease progression or death from any cause. OS was defined as time from EGFR TKI treatment initiation to death from any cause.
Subgroup analyses
We performed subgroup analyses to determine if the estimates of effects for the survival outcomes were influenced by: the type of EGFR TKI agents used (third- versus (vs) first- or second-generation), PD-L1 positivity threshold (≥ 1% vs H score ≥ 109.23 vs ≥ 5% vs ≥ 50%), type of antibody clones used (SP263 vs SP142 vs C288) and study design (retrospective analysis of RCT vs retrospective cohort study).
Statistical analysis
The log hazard ratios (HR) and their variances for time-to-event data (PFS and OS) were estimated using the published methods when appropriate summary statistics or Kaplan-Meier curves were reported [Citation19]. The individual trial log HR and their variances were combined using the generic inverse variance method. A HR of more than 1 indicates an advantage with PD-L1-negative tumors.
The restricted mean survival time (RMST) for both PFS and OS were estimated at the timepoints of 6 and 12 months from initiation of EGFR TKI treatment. We reconstructed the individual patient data (IPD) from the published Kaplan Meier curves using methods developed by Wei and colleagues [Citation20]. We estimated the restricted mean survival time using the methods developed by Cronin and colleagues [Citation21]. We calculated the differences in RMST and its standard deviation between PD-L1-positive and PD-L1-negative tumors at the prespecified timepoints. The individual study differences in RMST and their standard deviations were combined using the generic inverse variance method. A mean difference of less than zero indicates an advantage with PD-L1-negative tumors.
The chi-square Cochrane Q test was used to detect any heterogeneity across the different studies and between subgroups [Citation22,Citation23]. A p-value (p) of less than .05 would indicate a statistically significant difference between the groups, for example, a statistically significant heterogeneity among the studies’ results. The I squared (I2) statistics was used to judge the magnitude of heterogeneity. An I2 statistic of more than 25% would signify that at least moderate level of heterogeneity is present among the study results. The random-effect meta-analysis model was used in the analysis.
Statistical analysis was performed using Stata version 16.0 (Statacorp, Texas) and Cochrane RevMan version 5.3 (Cochrane Collaboration).
Results
Result of search strategy
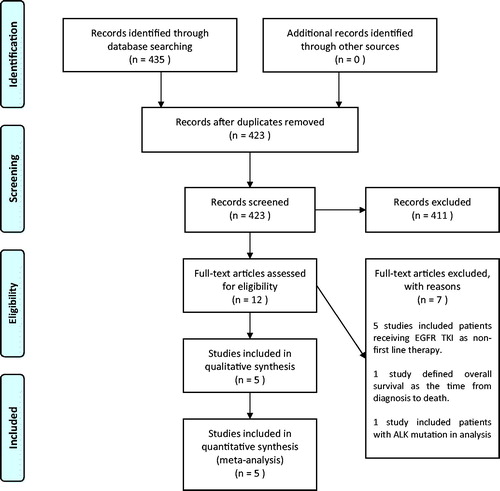
We identified 423 records from our search strategy (). After screening through the titles and abstracts, we retrieved the full text articles of twelve records for further evaluation. We excluded seven studies as these studies included patients receiving EGFR TKI as second line of treatment, defined OS from time of diagnosis or included patients with anaplastic lymphoma kinase (ALK) mutation in the analysis. Eventually, we included a total of five non-randomized comparative studies in the meta-analysis [Citation5,Citation24–27].
Figure 1. Flow diagram of study selection. ALK: anaplastic lymphoma kinase; EGFR: epidermal growth factor receptor; TKI: tyrosine kinase inhibitor.
These five studies met the prespecified eligibility criteria. Brown et al. performed a retrospective analysis of a phase III RCT, FLAURA trial, which randomized advanced EGFR-mutated NSCLC patients to receive either Osimertinib or Erlotinib/Gefitinib, and assessed whether PD-L1 expression is an effect modifier on PFS outcome in 106 patients [Citation5]. Hsu et al. conducted a retrospective observational study of 123 patients with advanced EGFR-mutated lung adenocarcinoma who developed primary resistance to EGFR-TKIs and reported the relationship between different PD-L1 expression levels and PFS and OS outcomes [Citation24]. Matsumoto et al. retrospectively investigated the efficacy of EGFR TKIs in 52 patients with advanced EGFR-mutated NSCLC according to the tumor microenvironment, based on the tumor expression of PD-L1 and CD8+ tumor-infiltrating lymphocytes [Citation25]. Soo et al. characterized the expression patterns of several immunoregulatory proteins including PD-L1 in 70 NSCLC patients with EGFR mutation and reported their prognostic significance in term of the associations with PFS outcome [Citation26]. Su et al. performed a retrospective cohort study including 84 eligible patients with advanced EGFR-mutated NSCLC which assessed the predictive effect of PD-L1 expression (strongly positive, weakly positive and negative) on the treatment response of EGFR TKI with regards to objective response rate and PFS [Citation27].
Characteristic of eligible studies
The characteristics of eligible studies were summarized in . One of the five included studies was a retrospective analysis of a RCT [Citation5]; while the remaining four studies were retrospective cohort studies [Citation24–27]. The median sample size of the included studies was 84 (range, 52–123). Four studies used first- or second-generation EGFR TKI treatment [Citation24–27]. Two studies set their PD-L1 positivity threshold as 1% [Citation5,Citation24], while the others used 5% [Citation27], 50% [Citation25] or H score of 109.23 [Citation26]. Two studies used SP263 [Citation5,Citation24] and two studies used SP142 antibody clones for PD-L1 testing [Citation26,Citation27].
Table 1. Characteristics of included studies.
The formal critical appraisal of the five studies indicated that the overall risk of bias in the methodologic quality was moderate ().
Table 2. Methodologic quality assessment of individual studies using ROBINS-I tool.
Progression-free survival
Brown et al.’s study provided two Kaplan-Meier curves demonstrating the PFS of patients in both Osimertinib and Erlotinib/Gefitinib treatment arms by the PD-L1 expression status (positive vs negative) [Citation5]. We reconstructed the time-to-event data using the published IPD method [Citation20] to enable the comparison of PFS between PD-L1-positive and PD-L1-negative patients in Osimertinib and Erlotinib/Gefitinib arms separately. Su et al.’s study provided a Kaplan-Meier curve showing the PFS of three groups of patients with strongly-positive, weakly-positive and negative PD-L1 expressions [Citation27]. We reconstructed the time-to-event data using the similar method by combining the patients with strongly-positive and weakly-positive PD-L1 expressions into one PD-L1-positive cohort. The other three studies provided Kaplan-Meier curves that have plotted the time-to-event data according to the dichotomous groups based on the PD-L1 expression status (PD-L1-positive and PD-L1-negative) [Citation24–26].
PD-L1-positive tumors were associated with clinically substantial and statistically significant higher hazards for disease progression or death compared to PD-L1-negative tumors (HR: 2.41; 95% confidence interval (CI): 1.59 to 3.66; p < .001) ().
Figure 2. (A) Pooled hazard ratios of progression-free survival in patients with stage IV EGFR-mutated NSCLC treated with first-line EGFR TKIs; (B) Pooled weight mean differences in restricted mean progression-free survival time at 6 and (C) 12 months since EGFR TKI treatment initiation. CI: confidence interval; EGFR: epidermal growth factor receptor; NSCLC: non-small cell lung cancer; PD-L1: programmed cell-death ligand 1; SD: standard deviation; TKI: tyrosine kinase inhibitor.
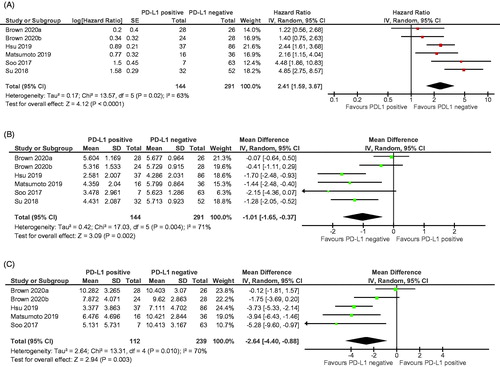
PD-L1-positive tumors had a significantly shorter restricted mean PFS time at the timepoints of 6 (WMD in the restricted mean PFS time (RMPFST6): −1.01; 95% CI: −1.65 to −0.37; p = .002) and 12 months (WMD in the RMPFST12: −2.64; 95% CI: −4.40 to −0.88; p = .003) compared to PD-L1-negative tumors (; Supplementary Data Citation2).
There was statistically significant heterogeneity in the HRs for PFS (I2 = 63.2%; chi-square p = .018) as well as for the WMD in the RMPFST6 (I2 = 70.6%; chi-square p = .004) and RMPFST12 (I2 = 70.0%; chi-square p = .010) among the individual studies ().
The GRADE quality of the summarized evidence for PFS outcomes was judged to be low to very low (Supplementary Data Citation3).
Subgroup analyses
The results of subgroup analyses for PFS were summarized in .
Table 3. Subgroup analyses.
The effect of PD-L1 expression on PFS outcomes was greater for studies using first- or second-generation rather than third-generation EGFR TKI, in terms of HR for PFS (HR, 2.69 vs 1.22; chi-square p = .069), RMPFST6 (WMD, −1.23 vs −0.07; chi-square p = .005) and RMPFST12 (WMD, −3.29 vs −0.12; chi-square p = .003).
There was significant effect modification by PD-L1 positivity threshold defined by four different positive thresholds on the HR for PFS (HR 4.85 (≥ 5%) vs 4.46 (H score ≥ 109.23) vs 2.16 (≥ 50%) vs 1.76 (≥ 1%), chi-square p = .018). There was no significant effect modification by PD-L1 positivity threshold on RMPFST6 and RMPFST12 outcomes.
There was significant effect modification by the type of antibody clones defined by three different clones on the HR for PFS (HR 4.73 (SP142) vs 2.16 (C288) vs 1.76 (SP263), chi-square p = .007). There was no significant effect modification by type of antibody clones on RMPFST6 and RMPFST12 outcomes.
The effect of PD-L1 expression on the PFS outcomes was greater for retrospective cohort studies compared to retrospective analysis of RCT, in terms of HR for PFS (HR, 3.11 vs 1.33; chi-square p = .005), RMPFST6 (WMD, −1.51 vs −0.21; chi-square p < .001) and RMPFST12 (WMD, −3.93 vs −0.86; chi-square p = .003).
Overall survival
Hsu et al.’s study was the only study that reported OS outcome [Citation24].
PD-L1-positive tumors were associated with clinically substantial and statistically significant higher hazards of death compared to PD-L1-negative tumors (HR, 2.28; 95% CI: 1.44–3.61; p < .001).
PD-L1-positive tumors had a significantly shorter restricted mean OS time at the timepoints of 12 months (WMD in the restricted mean OS time at 12 months (RMOST12), −1.55; 95% CI, −3.05 to −0.04; p = .044) and 24 months (WMD in the RMOST24, −5.04; 95% CI, −8.48 to −01.61; p = .004) but not at 6 months compared to PD-L1-negative tumors.
The GRADE quality of the summarized evidence for OS outcomes was judged to be low to very low (Supplementary Data 3).
Discussion
This systematic review and meta-analysis demonstrated that PD-L1-positive tumors were associated with significantly higher hazards for disease progression or death compared to PD-L1-negative tumors in stage IV EGFR-mutated NSCLC treated with first-line EGFR TKIs. However, the quality of evidence supporting this observation was judged to be low to very low.
A previous meta-analysis of six retrospective studies by Bai and colleagues found that positive PD-L1 expression was not significantly associated with PFS and OS in patients with advanced EGFR-mutated NSCLC receiving EGFR TKIs. This is not consistent with our study findings likely because Bai and colleagues included articles which evaluated EGFR TKIs as the second or subsequent lines of treatment [Citation28,Citation29]. The exposure to the other previous systemic treatments including chemotherapy and immunotherapy may have altered the PD-L1 expression characteristics of the original tumors and thus it will be difficult to establish an association between PD-L1 expression and efficacy of EGFR TKIs [Citation30–32].
There are several possible biological explanations for the lower efficacy of EGFR TKIs in PD-L1 positive, EGFR-mutated tumors. Firstly, high PD-L1 expression indicates evasion of tumor immune surveillance, especially in EGFR-mutated tumors [Citation33,Citation34]. Secondly, PD-L1 expression also reflects the immunogenicity of the tumor microenvironment, which was linked to high tumor mutational burden and aberrations of multiple oncogenic pathways [Citation35]. Thirdly, PD-L1 promotes the epithelial-mesenchymal transition in NSCLC which is a key process that drives tumor metastasis and aggressiveness [Citation35]. Fourthly, PD-L1 expression has been reportedly linked to TKI resistance via various regulatory mechanisms. For example, Lin et al. showed that PD-L1-mediated TKI resistance may occur as a result of persistent activation of extracellular signal-regulated kinase (ERK) signaling due to increased BIM phosphorylation and degradation via PD-L1/Bcl-2-associated athanogene-1 (BAG-1) axis [Citation36]. Tung et al. demonstrated that PD-L1 might be responsible for the upregulation of YAP1 which promotes immune escape of tumor cells and thus results in EGFR TKI resistance [Citation7]. In addition, a study by Peng et al. showed that the three common mechanism of acquired EGFR TKI resistance, namely c-MET amplification, hepatocyte growth factor (HGF) and T790M, were linked to upregulation of PD-L1 expression through different pathways including MAPK, PI3K and NF-kappa B pathways [Citation8].
Our subgroup analysis found that the effect of PD-L1 expression on PFS outcome was significantly smaller when third-generation EGFR TKI (Osimertinib) was used, comparing to older-generation EGFR TKIs. There are two possible explanations. Firstly, Osimertinib has a higher efficacy compared to Geftinib or Erlotinib for both PD-L1-positive and negative tumors. A landmark phase III RCT (FLAURA trial) demonstrated that Osimertinib significantly improved PFS and OS of patients with untreated advanced EGFR-mutated NSCLC compared to older-generation EGFR TKIs (Gefitinib or Erlotinib) [Citation37,Citation38]. Secondly, the sample size of patient population receiving Osimertinib was small, thus giving rise to a wide confidence interval.
Our study has several strengths. Firstly, we have strict definitions on our study eligibility criteria. For instance, we focused on studies that included solely patients with newly-diagnosed stage IV EGFR-mutated NSCLC treated with first-line EGFR TKIs in order to avoid the confounding effect of prior systemic treatment exposure. The selected studies must also define the PFS and OS outcomes from the start of treatment initiation rather than from time of diagnosis as the studies that estimated the effect of PD-L1 expression on the survival outcomes calculated from the time of diagnosis will incur immortal time bias. Secondly, we estimated RMST at prespecified timepoints in the analysis of survival outcomes to provide an absolute measure of the difference in survival times between PD-L1-positive and PD-L1-negative tumors. This would nicely complement the HRs estimated from the Cox regression analysis which represent a relative measure of the difference in survival outcomes between PD-L1-positive and PD-L1-negative tumors. Thirdly, we used validated instruments to judge the methodologic quality of the individual studies and the quality of the summarized evidence.
Our present study was limited by the non-randomized design of the included studies where the baseline characteristics could possibly be heterogeneous. Besides, our study was conducted based on the published data rather than individual patient data, thus limiting our ability to perform more granular analyses. While our subgroup analyses did not demonstrate any significant effect modification based on the PD-L1 positivity thresholds and type of antibody clones used for PD-L1 testing, it is recognized that there is a lack of standardization in IHC testing methods and result interpretation of PD-L1 expression [Citation39]. In addition, our study did not address the potential confounding effects from the presence of other concomitant mutations, such as TP53, RB1, MAP2K, MET amplification, ALK fusion and BRAF V600E, which might have strong prognostic influence [Citation40–42].
Our study findings have several implications on clinical practice. PD-L1 expression may be a potential negative prognostic biomarker in patients with advanced EGFR-mutated NSCLC receiving first-line EGFR TKIs. This is corresponding to the findings of other studies [Citation43,Citation44]. As patients with PD-L1-positive, EGFR-mutated NSCLC may have poorer prognosis, clinicians should consider exploring more aggressive treatment approach to optimize disease control and survival outcomes. For example, one may consider adding platinum-based doublet chemotherapy [Citation45,Citation46] or vascular endothelial growth factor (VEGF) receptor inhibitors (such as Bevacizumab or Ramucirumab) [Citation47,Citation48] in this population.
In conclusion, this systematic review provides the best available information on the impact of PD-L1 expression on the PFS outcomes in stage IV EGFR-mutated NSCLC treated with first-line EGFR TKIs. There is low certainty in the evidence to suggest that positive PD-L1 expression is associated with inferior disease control and survival outcomes in patients with stage IV EGFR-mutated NSCLC treated with first-line EGFR TKIs. More robust molecular studies on the co-expressed mutations and the tumor microenvironment and randomized trials are warranted to truly assess the prognostic and predictive values of baseline PD-L1 expression on EGFR-mutated NSCLC patients.
Author contributions
Supplemental Material
Download MS Word (26.7 KB)Disclosure statement
No potential conflict of interest was reported by the author(s).
References
- Ettinger DS, Wood DE, Aggarwal C, et al. NCCN guidelines insights: non-small cell lung cancer, Version 1.2020. J Natl Compr Canc Netw. 2019;17(12):1464–1472.
- Reck M, Rodriguez-Abreu D, Robinson AG, et al. Pembrolizumab versus chemotherapy for PD-L1-positive non-small-cell lung cancer. N Engl J Med. 2016;375(19):1823–1833.
- Mok TSK, Wu YL, Kudaba I, et al. Pembrolizumab versus chemotherapy for previously untreated, PD-L1-expressing, locally advanced or metastatic non-small-cell lung cancer (KEYNOTE-042): a randomised, open-label, controlled, phase 3 trial. Lancet. 2019;393(10183):1819–1830.
- Carbone DP, Reck M, Paz-Ares L, et al. First-line nivolumab in stage iv or recurrent non-small-cell lung cancer. N Engl J Med. 2017;376(25):2415–2426.
- Brown H, Vansteenkiste J, Nakagawa K, et al. Programmed cell death ligand 1 expression in untreated egfr mutated advanced nsclc and response to osimertinib versus comparator in FLAURA. J Thorac Oncol. 2020;15(1):138–143.
- Yoneshima Y, Ijichi K, Anai S, et al. PD-L1 expression in lung adenocarcinoma harboring EGFR mutations or ALK rearrangements. Lung Cancer. 2018;118:36–40.
- Tung JN, Lin PL, Wang YC, et al. PD-L1 confers resistance to EGFR mutation-independent tyrosine kinase inhibitors in non-small cell lung cancer via upregulation of YAP1 expression. Oncotarget. 2018;9(4):4637–4646.
- Peng S, Wang R, Zhang X, et al. EGFR-TKI resistance promotes immune escape in lung cancer via increased PD-L1 expression. Mol Cancer. 2019;18(1):165.
- D’Incecco A, Andreozzi M, Ludovini V, et al. PD-1 and PD-L1 expression in molecularly selected non-small-cell lung cancer patients. Br J Cancer. 2015;112(1):95–102.
- Tang Y, Fang W, Zhang Y, et al. The association between PD-L1 and EGFR status and the prognostic value of PD-L1 in advanced non-small cell lung cancer patients treated with EGFR-TKIs. Oncotarget. 2015;6(16):14209–14219.
- Soo RA, Lim SM, Syn NL, et al. Immune checkpoint inhibitors in epidermal growth factor receptor mutant non-small cell lung cancer: current controversies and future directions. Lung Cancer. 2018;115:12–20.
- Moher D, Liberati A, Tetzlaff J, et al. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. J Clin Epidemiol. 2009;62(10):1006–1012.
- Guo T, Ni J, Yang X, et al. Pattern of recurrence analysis in metastatic EGFR-mutant NSCLC treated with Osimertinib: implications for consolidative stereotactic body radiation therapy. Int J Radiat Oncol Biol Phys. 2020;107(1):62–71.
- Sterne JA, Hernan MA, Reeves BC, et al. ROBINS-I: a tool for assessing risk of bias in non-randomised studies of interventions. BMJ. 2016;355:i4919.
- Guyatt G, Oxman AD, Akl EA, et al. GRADE guidelines: 1. introduction-GRADE evidence profiles and summary of findings tables. J Clin Epidemiol. 2011;64(4):383–394.
- Sterne JAC, Savovic J, Page MJ, et al. RoB 2: a revised tool for assessing risk of bias in randomised trials. BMJ. 2019;366:l4898.
- Guyatt GH, Oxman AD, Kunz R, et al. What is “quality of evidence” and why is it important to clinicians? BMJ. 2008;336(7651):995–998.
- Guyatt GH, Oxman AD, Vist GE, et al. GRADE: an emerging consensus on rating quality of evidence and strength of recommendations. BMJ. 2008;336(7650):924–926.
- Tierney JF, Stewart LA, Ghersi D, et al. Practical methods for incorporating summary time-to-event data into meta-analysis. Trials. 2007;8:16.
- Wei Y, Royston P. Reconstructing time-to-event data from published Kaplan-Meier curves. Stata J. 2017;17(4):786–802.
- Cronin A, Tian L, Uno H. Strmst2 and Strmst2pw: new commands to compare survival curves using the restricted mean survival time. The Stata Journal. 2016;16(3):702–716.
- Higgins JP, Thompson SG. Quantifying heterogeneity in a meta-analysis. Stat Med. 2002;21(11):1539–1558.
- Higgins JPT, Thompson SG, Deeks JJ, et al. Measuring inconsistency in meta-analyses. BMJ. 2003;327(7414):557–560.
- Hsu KH, Huang YH, Tseng JS, et al. High PD-L1 expression correlates with primary resistance to EGFR-TKIs in treatment naïve advanced EGFR-mutant lung adenocarcinoma patients . Lung Cancer. 2019;127:37–43.
- Matsumoto Y, Sawa K, Fukui M, et al. Impact of tumor microenvironment on the efficacy of epidermal growth factor receptor-tyrosine kinase inhibitors in patients with EGFR-mutant non-small cell lung cancer. Cancer Sci. 2019;110(10):3244–3254.
- Soo RA, Kim HR, Asuncion BR, et al. Significance of immune checkpoint proteins in EGFR-mutant non-small cell lung cancer. Lung Cancer. 2017;105:17–22. Mar
- Su S, Dong ZY, Xie Z, et al. Strong programmed death ligand 1 expression predicts poor response and de novo resistance to EGFR tyrosine kinase inhibitors among NSCLC patients with EGFR mutation. J Thorac Oncol. 2018;13(11):1668–1675.
- Meniawy TM, Lake RA, McDonnell AM, et al. PD-L1 on peripheral blood T lymphocytes is prognostic in patients with non-small cell lung cancer (NSCLC) treated with EGFR inhibitors. Lung Cancer. 2016;93:9–16.
- Sorensen SF, Demuth C, Weber B, et al. Increase in soluble PD-1 is associated with prolonged survival in patients with advanced EGFR-mutated non-small cell lung cancer treated with Erlotinib. Lung Cancer. 2016;100:77–84.
- Song Z, Yu X, Zhang Y. Altered expression of programmed death-ligand 1 after neo-adjuvant chemotherapy in patients with lung squamous cell carcinoma. Lung Cancer. 2016;99:166–171.
- Guo L, Song P, Xue X, et al. Variation of programmed death ligand 1 expression after platinum-based neoadjuvant chemotherapy in lung cancer. J Immunother. 2019;42(6):215–220.
- Haratake N, Toyokawa G, Tagawa T, et al. Positive conversion of PD-L1 expression after treatments with chemotherapy and Nivolumab. Anticancer Res. 2017;37(10):5713–5717.
- Weisel K, Palumbo A, Chanan-Khan A, et al. Phase 3 randomised study of daratumumab, bortezomib and dexamethasone (DVd) vs bortezomib and dexamethasone (Vd) in patients (pts) with relapsed or refractory multiple myeloma (RRMM): CASTOR. Annals of Oncology. 2016;27:vi313. English.
- Santaniello A, Napolitano F, Servetto A, et al. Tumour microenvironment and immune evasion in EGFR addicted NSCLC: hurdles and possibilities. Cancers. 2019;11(10):1419.
- Lou Y, Diao L, Cuentas ER, et al. Epithelial-mesenchymal transition is associated with a distinct tumor microenvironment including elevation of inflammatory signals and multiple immune checkpoints in lung adenocarcinoma. Clin Cancer Res. 2016;22(14):3630–3642.
- Lin PL, Wu TC, Wu DW, et al. An increase in BAG-1 by PD-L1 confers resistance to tyrosine kinase inhibitor in non-small cell lung cancer via persistent activation of ERK signalling. Eur J Cancer. 2017;85:95–105.
- Soria JC, Ohe Y, Vansteenkiste J, et al. Osimertinib in untreated EGFR-mutated advanced non-small-cell lung cancer. N Engl J Med. 2018;378(2):113–125.
- Ramalingam SS, Vansteenkiste J, Planchard D, et al. Overall survival with Osimertinib in untreated, EGFR-mutated advanced NSCLC. N Engl J Med. 2020;382(1):41–50.
- Sholl LM. Programmed death ligand 1 immunohistochemistry: can we agree on this? Histopathology. 2020;76(2):189–190.
- Chen M, Xu Y, Zhao J, et al. Concurrent driver gene mutations as negative predictive factors in epidermal growth factor receptor-positive non-small cell lung cancer. EBioMedicine. 2019;42:304–310.
- Offin M, Chan JM, Tenet M, et al. Concurrent RB1 and TP53 alterations define a subset of EGFR-mutant lung cancers at risk for histologic transformation and inferior clinical outcomes. J Thorac Oncol. 2019;14(10):1784–1793.
- Aggarwal C, Davis CW, Mick R, et al. Influence of TP53 mutation on survival in patients with advanced EGFR-mutant non-small-cell lung cancer. J Cin Oncol Precis Oncol. 2018;2018(2):1–29.
- Kobayashi K, Seike M, Zou F, et al. Prognostic significance of NSCLC and response to EGFR-TKIs of EGFR-mutated NSCLC based on PD-L1 expression. Anticancer Res. 2018;38(2):753–762.
- Azuma K, Ota K, Kawahara A, et al. Association of PD-L1 overexpression with activating EGFR mutations in surgically resected nonsmall-cell lung cancer. Ann Oncol. 2014;25(10):1935–1940.
- Noronha V, Patil VM, Joshi A, et al. Gefitinib versus Gefitinib plus pemetrexed and carboplatin chemotherapy in EGFR-mutated lung cancer. J Cin Oncol. 2020;38(2):124–136.
- Hosomi Y, Morita S, Sugawara S, et al. Gefitinib alone versus Gefitinib plus chemotherapy for non-small-cell lung cancer with mutated epidermal growth factor receptor: NEJ009 study. J Cin Oncol. 2020;38(2):115–123.
- Saito H, Fukuhara T, Furuya N, et al. Erlotinib plus Bevacizumab versus Erlotinib alone in patients with EGFR-positive advanced non-squamous non-small-cell lung cancer (NEJ026): interim analysis of an open-label, randomised, multicentre, phase 3 trial. Lancet Oncol. 2019;20(5):625–635.
- Nakagawa K, Garon EB, Seto T, et al. Ramucirumab plus Erlotinib in patients with untreated, EGFR-mutated, advanced non-small-cell lung cancer (RELAY): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet Oncol. 2019;20(12):1655–1669.
