Abstract
Background
The widespread use of electronic patient-generated health data has led to unprecedented opportunities for automated extraction of clinical features from free-text medical notes. However, processing this rich resource of data for clinical and research purposes, depends on labor-intensive and potentially error-prone manual review. The aim of this study was to develop a natural language processing (NLP) algorithm for binary classification (single metastasis versus two or more metastases) in bone scintigraphy reports of patients undergoing surgery for bone metastases.
Material and methods
Bone scintigraphy reports of patients undergoing surgery for bone metastases were labeled each by three independent reviewers using a binary classification (single metastasis versus two or more metastases) to establish a ground truth. A stratified 80:20 split was used to develop and test an extreme-gradient boosting supervised machine learning NLP algorithm.
Results
A total of 704 free-text bone scintigraphy reports from 704 patients were included in this study and 617 (88%) had multiple bone metastases. In the independent test set (n = 141) not used for model development, the NLP algorithm achieved an 0.97 AUC-ROC (95% confidence interval [CI], 0.92–0.99) for classification of multiple bone metastases and an 0.99 AUC-PRC (95% CI, 0.99–0.99). At a threshold of 0.90, NLP algorithm correctly identified multiple bone metastases in 117 of the 124 who had multiple bone metastases in the testing cohort (sensitivity 0.94) and yielded 3 false positives (specificity 0.82). At the same threshold, the NLP algorithm had a positive predictive value of 0.97 and F1-score of 0.96.
Conclusions
NLP has the potential to automate clinical data extraction from free text radiology notes in orthopedics, thereby optimizing the speed, accuracy, and consistency of clinical chart review. Pending external validation, the NLP algorithm developed in this study may be implemented as a means to aid researchers in tackling large amounts of data.
Introduction
In medicine, electronic health record (EHR) data is increasing exponentially over time [Citation1]. The majority of this data is unstructured text in clinical reports, impeding its utilization in clinical practice and research setting. Manually extracting clinical characteristics of interest from these medical documents remain inefficient and prone to error; therefore neglecting potential valuable information [Citation2,Citation3]. One of these characteristics is the number of bone metastases as the quantity of bone metastases is associated with adverse outcomes such as postoperative complications and survival in oncologic populations [Citation4–6]. No diagnosis code or automated extraction tool is available to bypass error prone and time-consuming manual chart review.
Artificial intelligence (AI) has emerged as a powerful method to transform medical care [Citation7–9]. Although many AI-based methods have emerged in orthopedic healthcare with strong performance, analysis of free-text clinical notes remains challenging [Citation5]. One approach to analyze the free-text of patients’ medical records is the use of natural language processing (NLP), a subfield of AI that focuses on enabling computers to process human language [Citation10]. However, to our knowledge, there are no NLP algorithms available for extracting meaningful clinical features from free-text radiology reports in the field of orthopedic oncology.
The aim of this study was to develop an NLP algorithm for binary classification (single metastasis versus two or more metastases) in bone scintigraphy reports of patients undergoing surgery for bone metastases.
Material and methods
Guidelines
The TRIPOD guidelines were followed for the development of the algorithm reported in this study [Citation11].
Study population
Institutional review board approval was granted for retrospective review of EHRs. The inclusion criteria for this study were: (1) aged 18 years or older; (2) surgical treatment for a bone metastatic lesion; (3) date of procedure between 1 January 2002 and 1 January 2017; (4) index surgery at one of our two affiliated tertiary care hospitals; and (4) free-text bone scintigraphy reports within 6 months prior to the first index surgery in our institution’s EHR available for review. Metastatic lesions were accounted for in the axial or appendicular skeleton, and also included multiple myeloma and lymphoma [Citation12]. We excluded patients with (1) revision procedures, defined as any subsequent procedure after the index surgery addressing the metastatic lesion; and (2) kyphoplasty or vertebroplasty only. The selection criteria were based on previous published studies – in which ‘single versus multiple bone metastases’ a meaningful clinical feature was – that composed the cohort from which this current study extracted the bone scintigraphy reports. All patients in the cohort had at least a single bone metastasis. If a patient had multiple preoperative bone scintigraphy reports, the free-text report closest to surgery with a maximum of 6 months was obtained. If a patient underwent multiple surgeries, we considered the first surgery for bone metastases as the index procedure.
EHRs of patients in our institutional database of metastatic bone tumor were reviewed [Citation13,Citation14]. We identified 1780 potentially eligible patients after screening the medical records, of which 1076 patients did not have a preoperative bone scintigraphy within 6 months. A total of 704 radiology reports from 704 patients were included in this study ().
Ground truth
The primary outcome was defined as single versus multiple bone metastases. This was manually annotated from free-text bone scintigraphy reports using a binary classification (single metastasis versus two or more metastases). The 704 selected reports were manually reviewed by three independent research coordinators (NK, BPF, JK). Each reviewer was blinded to the labels generated by the other reviewers. No additional clinical information was provided beside the free-text bone scintigraphy reports. Conflicts between the three reviewers were resolved by final research fellows (OQG, MERB) to establish a ground truth. The accuracy for the three reviewers was calculated with the Cohen’s kappa as an interrater reliability estimate.
Statistics
Preprocessing
Prior analysis, the raw text notes required generic and approach-specific preprocessing steps. First, free-text reports were preprocessed in the following two ways: (1) cleaned from redundant or duplicate information (e.g., white spaces between paragraphs, time, date), line breaks, and stop words (e.g., ‘and’, ‘for’, ‘the’); and (2) stemming which reduces words into a common base or root (e.g., ‘increased’ and ‘uptake’ converted to ‘increas’ and ‘uptak’, respectively) (see Appendix, Supplemental Digital Content 1). This transformed the raw text into the most parsimonious representation of the lexical meaning in a text note. Second, the bag-of-words representation method was applied to describe the relative frequency of words within a free-text. In this method, a matrix is created with rows for every free-text notes and columns for words (tokens) in the bone scintigraphy notes that correspond with the occurrence and frequency of words in the scintigraphy notes. Third, the term frequency-inverse document frequency (TF-IDF) was used to adjust for common and very rare words. This method reflects how important a word is to a document and measures the number of times that words appear in a given document relative to the frequency of these words across all documents. The bag-of-words and TF-IDF were used as final input for the algorithm.
Data analysis
A stratified 80:20 split of the total dataset of 704 patients was done to create a training set (n = 563) and independent test set (n = 141). An extreme gradient boosting (XGBoost) machine learning algorithm was developed on the training set to detect multiple bone metastases [Citation15]. The final model was evaluated on the independent test set, which was not used in developing the NLP model. The output of the NLP model is binary classification (single vs multiple bone metastases). We used the following metrics to assess the model performance: (1) discrimination [area under the receiver operating curve (AUC), precision-recall curve (PRC), area under the precision-recall curve, sensitivity (recall), specificity, negative-predictive value (NPV), positive predictive value (PPV), F1-score, negative likelihood ratio (LLR-), positive likelihood ratio (LLR+)]; (2) calibration (calibration slope and intercept); and (3) overall performance (Brier score) [Citation16]. The Brier score ranges from 0 (perfect prediction) to 1 (worst prediction). For correct interpretation of the Brier score a comparison should be performed with the null-model Brier score, which assigns a predicted probability equal to the observed prevalence of the outcome to each patient – in this study the prevalence of multiple bone metastases in the dataset. A Brier score lower than the null model Brier score indicates greater performance of the algorithm (see Appendix, Supplemental Digital Content 2).
Local explanations were provided to enable the ability to highlight individuals words used by the algorithm to determine single versus multiple bone metastases in individual free-text scintigraphy reports [Citation17]. This figure will show features in green that increased the estimation of the likelihood of multiple metastases whereas the features in red are those that decreased the estimation of the likelihood of single metastases. Anaconda Distribution (Anaconda, Inc., Austin, Texas), Python (Python Software Foundation, Wilmington, Delaware), R version (The R Foundation, Vienna, Austria), and RStudio (RStudio, Boston, Massachusetts) were used for data analysis.
Results
A total of 704 free-text bone scintigraphy reports from 704 patients were included in this study and 617 (88%) had multiple bone metastases. The patients had a mean age of 62 (standard deviation of 12) and 374 (53%) were female. The interrater reliability was adequate; the three reviewers generally agreed with each other (kappa = 0.8). In the independent test set (n = 141) not used for model development, the NLP algorithm achieved AUC-ROC of 0.97 (), AUC-PRC of 0.99 (), calibration intercept of −0.41, and calibration slope of 0.73 for classification of single versus multiple bone metastases (). The Brier score for multiple bone metastases was 0.05 compared to the null model Brier score (score for algorithm that estimates a probability equal to the population prevalence of multiple metastases for every patient) of 0.011.
Figure 2. (A) Receiver operating curve and (B) Precision-Recall curves of NLP algorithm for multiple bone metastases in the independent testing set, n = 141. NLP: natural language processing.
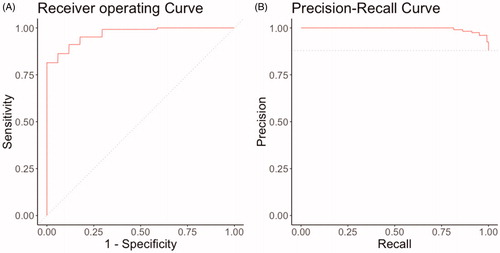
Table 1. Overall performance (95% confidence interval) of NLP algorithm for multiple bone metastases in the independent testing set, n = 141.
At a threshold of 0.10 and 0.90, the algorithm achieved a F1-score of 0.96 and 0.96, sensitivity of 0.99 and 0.94, specificity of 0.41 and 0.82, NPV of 0.88 and 0.67, and PPV of 0.92 and 0.97, respectively (). The algorithm, at the thresholds of 0.10 and 0.90, correctly classified the presence of multiple bone metastases in 123 and 117 reports (true positives) of the 124 who had multiple bone metastases in the testing cohort (sensitivity 0.99 and 0.94, respectively) and yielded 10 and 3 false positives (specificity 0.41 and 0.82, respectively). Local explanation of an actual free-text report demonstrated the specific words that drive toward (green) and against (red) classifying this report as a multiple bone metastasis (); the algorithm used words such as ‘increas’, ‘fractur’, and ‘active’ in the note to detect the occurrence of multiple bone metastases.
Figure 3. Example of local explanation at the individual patient-level explanation for multiple bone metastases. By color-coding the algorithm visualizes which words influence the prediction positively (green) or negatively (red) toward the outcome, in this case the presence of multiple bone metastases. In addition, the algorithm provides a prediction percentage, and depending on the chosen threshold by the user, the algorithm generates a labeling of the outcome (depicted at the bottom).

Table 2. Performance (95% confidence interval) of NLP algorithm at various thresholds for multiple bone metastases in the independent testing set, n = 141.
Discussion
Many clinical features have no procedural or diagnosis code, making them subject to error prone and labor-intensive manual chart review. The amount of bone metastases is a characteristic that lacks these codes but is associated with adverse outcomes such as postoperative complications and survival in oncologic populations [Citation4–6]. NLP constitutes a subfield of AI which shows promising results in analyzing the free-text included in EHRs [Citation10,Citation18,Citation19]. The goal of this study was to develop an NLP algorithm for the binary classification of single and multiple bone metastases in bone scintigraphy reports of patients undergoing surgery for bone metastases. Our NLP algorithm correctly classified the presence of multiple bone metastases in 117 of the 124 (sensitivity 0.94) who had multiple bone metastases in the testing cohort and yielded only 3 false positives (specificity 0.82). Pending external validation, the NLP algorithm developed in this study may be implemented as a means to aid clinicians and researchers in tackling large amounts of data.
The manual process of extracting clinical features from free-text can be time-consuming and labor-intensive, and can therefore produce variable results [Citation3]. With the recent widespread use of electronic medical records, the use of automated data extraction is on the rise [Citation1]. However, few studies used NLP to explore classification analysis of free-text radiology reports for patients with metastases as well as other malignancies. Senders et al. previously used NLP to quantify brain metastases in magnetic resonance imaging reports [Citation20]. Similarly, their NLP model had a high AUC of 0.92 and accuracy of 82%. Other NLP studies analyzing non-orthopedic oncologic radiology notes report comparable high AUCs ranging from 0.91 to 0.99 [Citation21–27]. In accordance with these studies, with a modest dataset (n = 1000), an NLP algorithm can be developed that extracts clinical features from free-text radiology notes. Compared to prior studies, this study developed algorithms capable of providing both estimations for likelihood of multiple bone metastases as well as explanations at the population and individual report level for multiple bone metastases.
The acceptability of a NLP algorithm’s error rate depends on the application. For example, if the intention in research is to accelerate the efficiency of manual review, higher false positive errors rates are less concerning. The ‘loss’ would be a reduced efficiency by increasing the number of charts reviewed. In clinical practice different error rates and evaluation metrics are important. For instance, achieving an <15% error rate in medical concept classification corresponds with human agreement on the same task [Citation28]; however, the error tolerance in daily practice might be lower, such as in misclassifying history of allergies or comorbidities. When developing a NLP algorithm, the tradeoffs between performance metrics have implications on potential biases and should be guided by the nature of the NLP task [Citation29].
We believe the NLP methods presented in this study may be useful in a range of orthopedics areas. First, a robust NLP tool could support research by rapidly identifying specific patients or diseases based on radiographical, pathological, or clinical findings. For example, creating a cohort of patients’ multiple bone metastases can propel research in understanding the impact of skeletal related events in this complicated patient population. In addition, the clinical feature ‘single versus multiple bone metastases’ can be used in various studies as a risk factor for an outcome, as was the case for the studies that supplied the bone-scintigraphy reports [Citation13,Citation14]. This could substantially reduce reviewer burden and error rate. Second, incorporating these NLP algorithms in EHRs may benefit population-based surveillance efforts. Third, NLP algorithms can be tailored for specific study designs; for example, the NLP algorithm developed in this study can extract clinical features that do not have specific administrative procedural or diagnosis code, such as the outcome in this study. Fourth, NLP algorithms can be used to ‘screen’ radiology reports for important information that may have been inadvertently missed by clinicians in daily practice. However, in view of the variability and complexity of used language in radiology reports, together with an imperfect NLP model, we believe that this NLP algorithm currently remains to be restricted for research purposes.
This study has limitations. First, this was a retrospective study with clinical notes from tertiary hospitals from one health-care system. Multi-institutional cohorts and prospective, temporal, and external validation of the NLP algorithm remains to be conducted to support generalizability of the study findings to other medical institutions. Nevertheless, this study provides a framework and supports an innovative approach for developing NLP models for automating the analysis of free-text radiology notes. Second, the ground truth for binary classification was manual review. Despite being labeled by three independent reviewers, human classification remains prone to error [Citation2,Citation3]. However, using human consensus in establishing the ground truth is a commonly used method in the absence of an absolute ground truth [Citation30]. Third, the NLP model was designed to classify single and multiple metastases in only bone scintigraphy reports. We did not design algorithms that would differentiate specific anatomic locations in reports of differing radiologic modalities. Future studies should incorporate the performance of NLP in non-bone scintigraphy radiology reports to quantify possible bone metastases and focus on differentiating the anatomical locations of bone metastases. Fourth, local explanation of the NLP algorithm identified some features (such as ‘fracture’ or ‘evid’) that appear to be clinically irrelevant to the presence of the bone metastases. Fracture may be clinically relevant because patients who had a pathologic fracture are more likely to have disseminated/advanced disease with multiple bone metastases. Words/tokens like ‘evid’ may represent the features of radiologist lexicon when delineating multiple metastases in our cohort but may represent overfitting to the available data such that the models are not transportable to new, independent data. Moreover, although over 50 radiologists contributed to this dataset from two different hospitals, all radiology reports were from one health-care system with potentially use of fixed phrases to express certain type of findings. The algorithm may make accurate predictions in this study sample but may not generalize to other datasets. This emphasizes the need for external validation of the study findings in order to support generalizability of the NLP algorithm to other medical institutions. Fifth, future research may include other machine learning-based NLP algorithms such as convolutional and recurrent neural networks that may improve the performance demonstrated here. Sixth, over half of the patients were excluded due to the two exclusion criteria from this current study design. Comparing baseline characteristics demonstrated several differences between the included and excluded groups (see Appendix, Supplemental Digital Content 3). However, these clinical differences are not relevant for this study since it has no implications on the study aim or the developed NLP model as the model does not take into account clinical, demographic, diagnosis, or treatment characteristics. Nevertheless, we deem the limitations proportionate to the strength of this NLP study. This study provides a proof-of-concept of applying similar NLP techniques to extract clinical features without procedural or diagnosis codes. To our knowledge, this is the first NLP study assessing an NLP algorithm for extracting clinical features without medical codes from free-text bone scintigraphy reports in the field of orthopedic oncology. By using thorough crosschecked manual labeling, this study provides valuable insights into the use of NLP in in orthopedics and its future role in clinical and research setting.
In conclusion, the widespread use of electronic patient-generated health data has led to unprecedented opportunities for research purposes. AI-based NLP methods enable us to automate the transformation of these unstructured free-text to clinical features, thereby optimizing the speed, accuracy, and consistency of clinical chart review. This study provides an NLP algorithm that has the potential to automate clinical data extraction from radiology notes in orthopedics. Pending external validation, the NLP algorithm developed in this study may be implemented as a means to aid clinicians and researchers in tackling large amounts of data.
Author contributions
OQG and MERB contributed to the research design and drafting the paper. AVK contributed to analyzing the data. NDK, BPF and JK contributed to processing and labeling the data. JJV and JHS contributed to revising the finalized paper critically.
Investigation performed at Massachusetts General Hospital
Supplemental Material
Download MS Word (15.8 KB)Supplemental Material
Download MS Word (14.9 KB)Supplemental Material
Download MS Word (14.1 KB)Acknowledgments
The authors thank D. Hayden, PhD from Harvard Catalyst Biostatistics Consultation for his help in the design and statistics.
Disclosure statement
Each author certifies that he or she has no commercial associations (e.g., consultancies, stock ownership, equity interest, patent/licensing arrangements, etc.) that might pose a conflict of interest in connection with the submitted article.
All ICMJE Conflict of Interest Forms for authors and Acta Oncologica® editors and board members are on file with the publication and can be viewed on request.
Additional information
Funding
References
- Peterson ED. Machine learning, predictive analytics, and clinical practice: can the past inform the present? J Am Med Assoc. 2019;322(23):2283–2284.
- Mi MY, Collins JE, Lerner V, et al. Reliability of medical record abstraction by non-physicians for orthopedic research. BMC Musculoskelet Disord. 2013;14:181.
- Cruz CO, Meshberg EB, Shofer FS, et al. Interrater reliability and accuracy of clinicians and trained research assistants performing prospective data collection in emergency department patients with potential acute coronary syndrome. Ann Emerg Med. 2009;54(1):1–7.
- Paulino Pereira NR, Ogink PT, Groot OQ, et al. Complications and reoperations after surgery for 647 patients with spine metastatic disease. Spine J. 2019;19(1):144–156.
- Karhade AV, Thio QCBS, Ogink PT, et al. Predicting 90-Day and 1-year mortality in spinal metastatic disease: development and internal validation. Neurosurgery. 2019;85(4):E671–7.
- Janssen SJ, Kortlever JTP, Ready JE, et al. Complications after surgical management of proximal femoral metastasis: a retrospective study of 417 patients. J Am Acad Orthop Surg. 2016;24(7):483–494.
- Obermeyer Z, Emanuel EJ. Predicting the future-big data, machine learning, and clinical medicine. N Engl J Med. 2016;375(13):1216–1219.
- Rajkomar A, Dean J, Kohane I. Machine learning in medicine. N Engl J Med. 2019;380(14):1347–1358.
- Wallis C. How artificial intelligence will change medicine. Nature Research. 2019;576(7787):S48–S48.
- Nadkarni PM, Ohno-Machado L, Chapman WW. Natural language processing: an introduction. J Am Med Inform Assoc. 2011;18(5):544–551.
- Collins GS, Reitsma JB, Altman DG, et al. Transparent reporting of a multivariable prediction model for individual prognosis or diagnosis (TRIPOD): the TRIPOD Statement. BMC Med. 2015;13:1
- Nathan SS, Healey JH, Mellano D, et al. Survival in patients operated on for pathologic fracture: implications for end-of-life orthopedic care. J Cin Oncol. 2005;23(25):6072–6082.
- Groot OQ, Ogink PT, Paulino Pereira NR, et al. High risk of symptomatic venous thromboembolism after surgery for spine metastatic bone lesions: a retrospective study. Clin Orthop Relat Res. 2019;477(7):1674–1686.
- Groot OQ, Ogink PT, Janssen SJ, et al. High risk of venous thromboembolism after surgery for long bone metastases: a retrospective study of 682 patients. Clin Orthop Relat Res. 2018;476(10):2052–2061.
- Chen T, Guestrin C. Xgboost. A scalable tree boosting system. Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining. ACM; 2016. p. 785–794.
- Brier GW. Verification of forecasts expressed in terms of probability. Mon Wea Rev. 1950;78(1):1–3.
- Ribeiro MT, Singh S, Guestrin C. “Why should i trust you?: Explaining the predictions of any classifier.” Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining. ACM; 2016. p. 1135–1144.
- Liang H, Tsui BY, Ni H, et al. Evaluation and accurate diagnoses of pediatric diseases using artificial intelligence. Nat. Med. 2019;25(3):433–438.
- Karhade AV, Bongers MER, Groot OQ, et al. Natural language processing for automated detection of incidental durotomy. Spine J. 2020;20(5):695–700.
- Senders JT, Karhade AV, Cote DJ, et al. Natural language processing for automated quantification of brain metastases reported in free-text radiology reports. J Am Med Clin Cancer Inform. 2019;3:1–9.
- Chen P-H, Zafar H, Galperin-Aizenberg M, et al. Integrating natural language processing and machine learning algorithms to categorize oncologic response in radiology reports. J Digit Imaging. 2018;31(2):178–184.
- Bozkurt S, Gimenez F, Burnside ES, et al. Using automatically extracted information from mammography reports for decision-support. J Biomed Inform. 2016;62:224–231.
- Ping XO, Tseng YJ, Chung Y, et al. Information extraction for tracking liver cancer patients’ statuses: from mixture of clinical narrative report types. Telemed J E Health. 2013;19(9):704–710.
- Carrell DS, Halgrim S, Tran DT, et al. Using natural language processing to improve efficiency of manual chart abstraction in research: the case of breast cancer recurrence. Am J Epidemiol. 2014;179(6):749–758.
- Sippo DA, Warden GI, Andriole KP, et al. Automated extraction of BI-RADS final assessment categories from radiology reports with natural language processing. J Digit Imaging. 2013;26(5):989–994.
- Sevenster M, Bozeman J, Cowhy A, et al. Automatically pairing measured findings across narrative abdomen CT reports. Annu Symp Symp Proc. 2013;2013:1262–1271.
- Glaser AP, Jordan BJ, Cohen J, et al. Automated extraction of grade, stage, and quality information from transurethral resection of bladder tumor pathology reports using natural language processing. J Am Med Clin Cancer Inform. 2018;2:1–8.
- Uzuner Ö, South BR, Shen S, et al. 2010 i2b2/VA challenge on concepts, assertions, and relations in clinical text. J Am Med Inform Assoc. 2011;18(5):552–556.
- Kohane IS. Using electronic health records to drive discovery in disease genomics. Nat Rev Genet. 2011;12(6):417–428.
- Valizadegan H, Nguyen Q, Hauskrecht M. Learning classification models from multiple experts. J Biomed Inform. 2013;46(6):1125–1135.