Abstract
Background
This article reviews the current knowledge on circulating tumor DNA (ctDNA) in early stage colon cancer and ongoing trials on ctDNA-guided treatment in the adjuvant setting.
Methods
A literature search of Pubmed was performed to identify studies on ctDNA in early stage colon cancer and neoadjuvant or adjuvant treatment. For ongoing trials, we searched clinicaltrials.gov and the Australian New Zealand Clinical Trials Registry (ANZCTR).
Results
Several studies show that ctDNA is a strong predictor for recurrence and survival after surgery and adjuvant chemotherapy. The specificity of this marker is extremely high, and the sensitivity is increasing with the development of technology. Recurrences can be detected very early and the analysis can potentially be used to guide neoadjuvant and adjuvant treatment. Ongoing and planned studies are now looking into escalation and de-escalation of therapy according to ctDNA-status after surgery.
Conclusion
Serial measurement of ctDNA shows great promise as a marker for both prognosis and response to treatment in early colon cancer. Future studies will show whether we can use this analysis for tailoring treatment for patients in the adjuvant and neoadjuvant setting. With improved technology, ctDNA has the potential of becoming a ‘game-changer’ in the treatment of early stage colon cancers.
Background
Colon cancer is the third most common malignancy in Sweden and the incidence is rising. The mortality is, however, declining which may be due to more effective treatments over time [Citation1] and earlier stage of disease at diagnosis [Citation2]. Worldwide, colon cancer is the second most common cause of cancer death and the fourth most common cancer with over one million new cases per year [Citation3].
The principal treatment of early stage disease is still curative intent surgery followed by adjuvant chemotherapy (ACT) in patients with high risk of recurrence. Recently, neoadjuvant chemotherapy (NAC) followed by surgery has emerged as an alternative strategy to treat certain cases with localized colon cancer [Citation4].
Twenty to 30% of patients with early-stage colon cancer relapse despite surgical and oncological treatment. A few prognostic and predictive factors are known, but we need to improve our ability to select patients with high risk of recurrence for additional, tailored therapies and avoid overtreating cases that are most likely cured by surgery alone. Known prognostic factors for recurrence are clinical stage according to the TNM classification [Citation5] and pathological stage of the surgically removed tumor [Citation6]. The tumor marker carcinoembryonic antigen (CEA) may be used as a complement for risk assessment but has limited specificity and sensitivity [Citation7].
Another prognostic and possibly predictive factor in early colorectal cancer is tumor microsatellite instability (MSI), which is the result of a deficiency in the mismatch repair genes of the cell. Around 15–20% of patients with newly diagnosed colorectal cancers have sporadic or inherited, that is, Lynch syndrome, deficiency of the mismatch repair protein [Citation8–12].
Furthermore, there is evidence that an immunoscore-based approach with analysis of tumor CD3+ and cytotoxic CD8+ T lymphocyte densities, is associated with survival and response to chemotherapy in stage III colon cancer [Citation13].
A novel factor to take into consideration is the level of circulating fragments of DNA. All healthy individuals harbor cell-free DNA (cfDNA) in plasma which originates from apoptotic, necrotic and shedding from living cells. In patients with cancer, a fraction of the cfDNA originates from cancer cells, i.e., circulating tumor DNA (ctDNA) [Citation14,Citation15]. The levels of ctDNA at diagnosis vary greatly between different types of tumors, among individual patients and different stages of the disease. High levels can usually be found in patients with metastases and detectable levels after surgery indicate the presence of minimal residual disease (MRD) [Citation16]. Early reports from smaller prospective studies suggests a very high prognostic value of detectable postoperative ctDNA in patients with colon cancer [Citation17–20].
Circulating tumor DNA can be analyzed by a tumor-informed or panel-based method. The tumor-informed approach examines a certain number of mutations in the patient’s own tumor, whereas the panel-based method does not require knowledge of the individual patient’s mutations and can include alterations such as methylations.
In early stage colon cancer, there is, thus, a need to find better and more accurate ways to predict recurrence after surgery and assess response to ACT and NAC. Monitoring of ctDNA in the perioperative setting and during follow-up is emerging as a new and highly promising marker for prognosis as well as therapeutic efficacy. We herein review the current status of clinical trials and future perspectives of this exciting field in early colon cancer.
Methods
For this review, we performed a search of PubMed with the keywords ‘colon cancer’ or ‘colorectal cancer’ or ‘colonic neoplasm’ and ‘ctDNA’ or ‘circulating tumor DNA’ or ‘liquid biopsy’ and ‘adjuvant’ or ‘neoadjuvant’.
For information regarding ongoing trials, we searched the US National Library of Medicine in clinicaltrials.gov and the Australian New Zealand Clinical Trials Registry (ANZCTR), with the use of the keywords ‘colorectal cancer’, ‘colon cancer’, ‘ctDNA’ or ‘circulating tumor DNA’. Among the clinical trials, we choose to present trials on adjuvant or neoadjuvant treatment, which randomize participants to treatment according to ctDNA status. Information on the Dutch (MEDOCC-CrEATE), Japanese (CIRCULATE-Japan) and French/European (CIRCULATE-EUROPE) trials, has been obtained from the respective research group.
Results
Circulating tumor DNA as a prognostic and predictive marker in adjuvant treatment of Colon cancer
In colorectal cancer, ctDNA can be found at different levels before surgery, depending on tumor stage and sensitivity of the analysis. Levels of preoperative ctDNA vary from 63.8% to 88.5% in different studies [Citation17,Citation18]. Furthermore, several trials have demonstrated a strong influence of postoperative ctDNA status on survival [Citation17,Citation21,Citation22]. These trials differ in setting, size, outcome measure and analysis ().
Table 1. Results of trials on ctDNA as a prognostic and predictive marker in early stage colorectal cancer.
A Danish cohort study of 125 patients with colorectal cancer stage I–III, showed that patients with positive ctDNA were seven times more likely to relapse than ctDNA negative cases, hazard ratio (HR: 7.2). If ctDNA positiveness persisted after adjuvant chemotherapy, these patients were 17 times more likely to relapse than ctDNA negative cases (HR: 17.5) [Citation17].
Likewise, this strong association between recurrence and ctDNA was corroborated in a study of 58 patients in Swedish hospitals with colorectal cancer stage I–III [Citation21]. In this study, none of the 45 ctDNA-negative patients experienced relapse during a median follow-up of 49 months. Moreover, detection preceded clinical and radiological signs of recurrence by a median of 3 months in the 13 patients with positive ctDNA during follow-up.
An Australian study of 230 stage II colon cancer patients by Tie and coworkers, found that 20 out of 230 patients (8.7%) were ctDNA positive 4–10 weeks after surgery [Citation20]. These patients had a markedly worse 3-year recurrence-free survival (RFS) than those with negative ctDNA (HR: 18), with RFS 0% for ctDNA positive patients versus 90% for ctDNA-negative cases. Postoperative ctDNA positivity had a greater influence on RFS than any one clinicopathological factor or any combination of these factors. The sensitivity of postoperative ctDNA to predict recurrence at 3 years was 48% and the specificity was 100%.
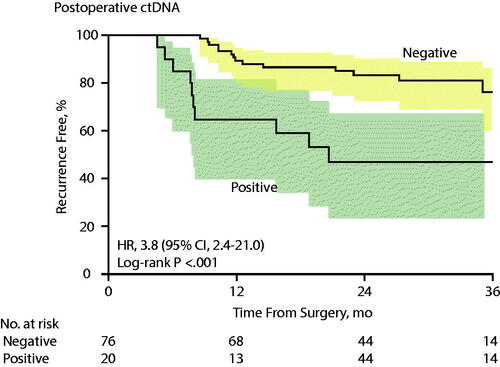
In yet another report by the Australian group, 96 patients with stage III colon cancer were studied and 21% were found to have detectable postoperative ctDNA [Citation19]. The 3-year RFI (recurrence-free interval) with positive ctDNA in the circulation was 47% versus 76% for those with negative ctDNA (). Furthermore, the samples taken after ACT were associated with a 3-year RFI of 30% for those with positive ctDNA and 77% for those with nonmeasurable levels of ctDNA (HR: 6.8).
Figure 1. Recurrence-Free Interval (RFI) in stage III colon cancer patients according to post-operative ctDNA, Kaplan-Meier Estimates (published with permission, Tie et al. JAMA Oncol 2019).
Several studies have also shown that repeated measurements of ctDNA after surgery in patients with colon cancer can detect recurrence earlier than repeated testing of CEA and radiology exams [Citation17–21]. However, all these trials have some methodological limitations, e.g., limited number of patients, discordancy of ctDNA sampling in relation to CT-scans and testing for CEA.
Furthermore, there are indications that repeated ctDNA measurements during treatment could be used as a predictive marker to guide therapy in colorectal cancer. Data from prospective studies with serial ctDNA measurements show that some patients can convert from ctDNA positivity to negativity after ACT, but if ctDNA remains positive, the recurrence risk is very high [Citation18,Citation20]. Moreover, the dynamics of ctDNA testing seem to give early indications of response and resistance to systemic therapy. This suggests an opportunity to intensify or change treatments in patients who remain ctDNA positive during ACT.
Ongoing and planned randomized clinical trials on ctDNA and adjuvant treatment
European trials
There are several newly started European trials on ctDNA as a predictor of therapeutic response and on escalation or de-escalation of ACT (). The large German/Austrian/Swiss CIRCULATE-AIO trial is an ongoing randomized adjuvant trial in patients with stage II microsatellite-stable (MSS) colorectal cancer ( and ) [Citation23]. Circulating tumor DNA is measured at inclusion and patients are grouped according to ctDNA status. Patients with positive postoperative ctDNA are randomized 2:1 to chemotherapy with investigator’s choice of chemotherapy: Capecitabine (Cape) for 6 months or Capecitabine and Oxaliplatin (CapOx) for 3 or 6 months, or follow-up. The ctDNA-negative patients are randomized 1:4 to follow-up within or outside the trial. The primary endpoint is disease-free survival (DFS).
Figure 2. Simplified description of design of the ongoing CIRCULATE-AIO adjuvant trial in localized colon cancer (n ≈ 4800).
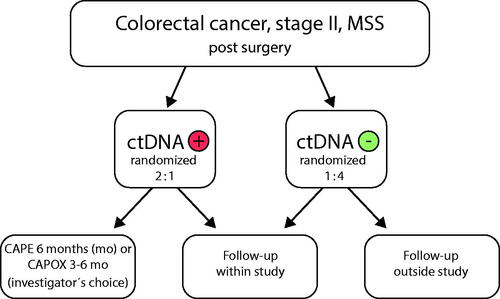
Table 2. Ongoing European adjuvant trials in early stage colon cancer evaluating the use of ctDNA.
The large French multicenter adjuvant trial, CIRCULATE 2-PRODIGE 70, has already included 300 stage II colon cancer patients, excluding T4b cases, and the researchers aim to recruit 2000 patients. Participants will follow a surveillance program according to current French guidelines and the 198 ctDNA positive patients will be randomized 2:1 to 6 months FOLFOX (5-FU and Oxaliplatin) or close follow-up () [Citation24,Citation25]. The primary endpoint is DFS.
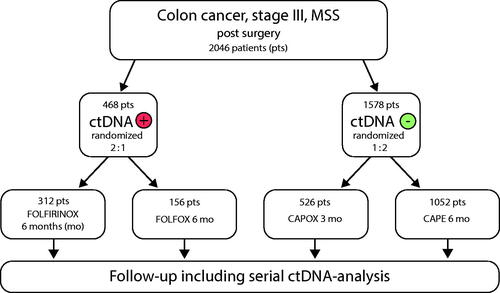
CIRCULATE 3-EUROPE is another large French randomized controlled adjuvant trial in its final stages of planning and approval (). Two thousand patients with stage-III colon cancer will be recruited to receive different ACTs according to ctDNA status. The ctDNA-positive patients are randomized to either FOLFIRINOX (5-FU, Irinotecan and Oxaliplatin) or FOLFOX for 6 months, whereas the ctDNA-negative group will receive either CapOx for 3 months or Cape for 6 months.
Figure 3. Simplified description of design of the proposed CIRCULATE 3-EUROPE adjuvant trial in early stage colon cancer (n = 2046).
Tracking Mutations in Cell Free Tumor DNA to Predict Relapse in Early Colorectal Cancer (TRACC) is a UK adjuvant trial. The investigators aim to include 1000 patients with high risk stage II or stage III colorectal cancer after R0 resection who have measurable ctDNA preoperatively () [Citation26,Citation27]. Patients are randomized to either standard of care (SOC) ACT or ctDNA guided ACT. In the guided arm, ctDNA negative cases receive de-escalated ACT for 3 months with single Cape, or no chemotherapy. Analysis of ctDNA is repeated at 3 months and if elevated, the patients are recommended the addition of 3 months CapOx. Investigators also aim to measure the incidence of positive ctDNA in this group of patients before surgery and examine the correlation between ctDNA levels and DFS postoperatively.
The researchers of the ongoing Danish phase II trial IMPROVE-IT study patients with ctDNA positive stage I-II colorectal cancer, who are not eligible for ACT according to Danish guidelines and who have a clear postoperative PET scan (). Patients with a clear PET scan are randomized to chemotherapy with CapOx (or FOLFOX) or intensified follow-up [Citation28]. Additionally, IMPROVE-IT2, is a randomized controlled trial where investigators look into ctDNA-guided surveillance as compared to standard surveillance with CT scans at 12 and 36 months after surgery according to Danish guidelines. The plan is to include 1800 patients and the primary endpoint is the number of patients with disease-recurrence who receive curative intended or local metastases-directed treatment [Citation29].
Additional trials include the Italian Pegasus trial [Citation30] and the Dutch MEDOCC-CrEATE study [Citation31] ().
The Swedish CITCCA pilot study [Citation32] has started recruitment in October 2020 and its purpose is to demonstrate the feasibility of prospective analysis of ctDNA in a cohort of 300–400 patients with colorectal cancer stage I–III. Our aim is to widely examine a great number of mutations for every patient in order to increase sensitivity. We will use a similar technique as Parsons and colleagues where tumor tissue will be sequenced using a broad pan-cancer targeted sequencing assay to enable MRD tracking, using a combination of mutations and structural variants [Citation33]. Patient-specific tests will then be designed and used for the detection of ctDNA. Sampling of ctDNA will occur preoperatively, 4–6 weeks after surgery, after 3 and 6 months and after 1 and 2 years. If the study confirms the results of previous studies and is successful in applying ctDNA profiling, we plan to initiate/participate in a large controlled trial, randomizing patients to adjuvant treatment according to ctDNA status.
Non-European trials
The Australian group has completed the recruitment of 459 patients in the DYNAMIC-II trial [Citation34] () which studies ctDNA guided ACT in stage II colon cancer versus SOC ACT. The primary outcome measure is RFS and the results of the trial are eagerly awaited. Patients randomized to arm A with positive ctDNA received ACT and those with negative ctDNA received no chemotherapy. Arm B patients were treated at their oncologist’s discretion and doctors were, at least initially, blinded to the ctDNA results. Accepted chemotherapy regimens were 3–6 months 5-FU or Cape or doublet treatment with Oxaliplatin.
Table 3. Ongoing non-European adjuvant trials in early stage colon cancer evaluating the use of ctDNA.
The DYNAMIC-III adjuvant trial [Citation35] in stage III colon cancer has also recruited well, i.e., almost 600 patients, and the investigators aim to include 1000 participants (). The design is complex and involves comparing SOC ACT versus ctDNA-guided treatment. Researchers aim to assess the effect on RFS of ctDNA-guided escalation and de-escalation of treatment.
The Australian ACT trial in rectal cancer, DYNAMIC-rectal, is also recruiting well and has so far included over 200 patients, with the target of 408 participants [Citation36]. Patients are randomized between SOC and ctDNA-guided treatment, where patients with a high-risk tumor and/or a ctDNA-positive result, receive chemotherapy with 4 months of Cape/5-FU with or without Oxaliplatin. The ctDNA-negative group will not receive ACT.
In the US, investigators will include 1400 patients with colon cancer stage IIA in the adjuvant COBRA trial (NRG-GI005) () [Citation37]. Arm A will undergo active surveillance and have blood stored for future ctDNA testing. Arm B will be tested for ctDNA at baseline and patients with positive ctDNA will receive ACT with FOLFOX for 6 months. Patients with negative ctDNA will undergo active surveillance. The researchers aim to examine ctDNA clearance after ACT and RFS for patients with positive ctDNA who have, or have not, received chemotherapy.
Moreover, CIRCULATE-US (NRG-GI008) is a trial where trialists, according to the preliminary design, will investigate ctDNA guided treatment in low risk, T1-3 N1, stage III colon cancer and plan to recruit around 1500 patients [Citation38]. Patients will be randomized to treatment according to postoperative ctDNA status. The ctDNA negative patients will be randomized to either CapOx/FOLFOX or serial measurements of ctDNA. If a participant in the latter group should develop a positive ctDNA result during follow-up, the patient is then randomized to delayed ACT with either CapOx/FOLFOX or mFOLFIRINOX for 6 months. Patients with a postoperative positive ctDNA will be randomized to either CapOx/FOLFOX or mFOLFIRINOX for 6 months. The primary endpoint is ctDNA status and DFS and the investigators plan to start recruitment in q3 2021.
Japanese researchers have initiated three parallel studies: GALAXY, VEGA and ALTAIR, which are included in a study platform called CIRCULATE-Japan [Citation39]. GALAXY is an observational study that includes monitoring of ctDNA in stage I-IV colorectal cancer. VEGA (), is a randomized phase-III trial where high-risk stage II and low-risk stage III colon cancer patients who are ctDNA negative after surgery, are randomized to either no chemotherapy or CapOx for 3 months. The primary endpoint is DFS. Finally, ALTAIR is a randomized, double-blind phase-III study where patients with colorectal cancer stage II–III who are positive for ctDNA after surgery, receive either placebo or 6 months of Trifluridine/Tipiracil. In total, these studies will include 2500 patients.
Additionally, the US SU2C randomized phase-III trial will answer the question whether early treatment in ctDNA positive disease after ACT is of benefit [Citation38,Citation39]. Investigators plan to recruit around 500 patients with stage III colon cancer, measuring ctDNA after ACT. Patients with positive ctDNA will be randomized to either 5-FU and Irinotecan (FOLFIRI) or observation whereas ctDNA negative patients will be observed. A subgroup of ctDNA positive, MSI-H patients will receive immunotherapy with Nivolumab. Primary endpoints are DFS in ctDNA positive patients and ctDNA clearance.
Discussion and future perspectives
Serial measurement of ctDNA has emerged as a highly promising method to detect MRD after surgery in patients with early stage colon cancer and to guide ACT in real-time. Several studies have shown excellent specificity and increasing sensitivity for ctDNA detection.
There are, however, several challenges in the detection of ctDNA.
Firstly, the cfDNA concentration in plasma is low, typically <10 ng/ml which represents ∼3000 genome copies. Furthermore, the fraction of ctDNA in cfDNA is low in localized cancer, commonly 0.01−0.0001, which requires advanced technologies for detection [Citation41]. Thus, the efficiency has to be high to minimize the loss of genome copies in the laboratory process to, in turn, maximize sensitivity in relation to the limited starting volume of blood. However, the low biological signal also requires high specificity, below the technical error rates of conventional high throughput sequencing technologies. This has spurred the introduction of molecular barcodes whilst preparing the DNA for sequencing [Citation42]. The use of molecular barcodes make it possible to determine which DNA molecules originate from the same original DNA molecule after polymerase chain reaction (PCR). This, in turn, allows for error correction and lowering of the technical noise below the biological signal [Citation41].
The half-life of ctDNA in patients with colon cancer is short, that is, 114 min [Citation43], which allows for real-time analysis. While multiple technologies may be adopted for detection of ctDNA in localized cancer such as beaming or digital droplet PCR [Citation33,Citation44], these are commonly limited to a pre-defined set of somatic alterations at certain positions [Citation45]. High-throughput sequencing, on the other hand, may be adopted to interrogate all somatic variants detected from tumor tissue, thereby increasing the sensitivity with several orders of magnitude [Citation33].
Several independent research groups have detected MRD in patients with colon cancer with the use of high-throughput sequencing. However, the number of genes analyzed differ, which is reflected in the reported variable degrees of sensitivity [Citation17,Citation20,Citation21]. Furthermore, cfDNA preserves the epigenetic imprint of the originating tissue [Citation46]. Recently reported data indicate that assessing the methylation status of informative regions may be applied for early detection of localized cancer, including colorectal cancer. This may enable MRD-assessment without a priori knowledge of the mutation status [Citation47,Citation48]. However, high-dimensional epigenetic profiling requires large training and validation sets to establish assay properties and potentially clinical utility. It is not clear, at this stage, how such assays perform compared to tracking somatic alterations.
In our CITCCA pilot trial [Citation32], we plan to further improve sensitivity by broad genomic profiling which we hope will perform better than techniques used in previous studies.
Currently, many studies are exploring the concept of escalation and de-escalation of ACT according to ctDNA status ( and ). The results of these studies will deepen our understanding of this approach and hopefully lead to more informed treatment decision-making in patients with early colon cancer.
After the initial results of the FOxTROT study on NAC in early colon cancer [Citation4], we may also have the alternative to recommend neoadjuvant chemotherapy prior to surgery in a selected group of high-risk patients. The preliminary results show that 6 weeks of NAC followed by surgery lead to a reduction in incomplete surgical resections. Results on the primary end-point, DFS at 2 years, are expected to be updated in the near future. The FOxTROT data probably need to be considered in the planning of upcoming trials on ctDNA in early colon cancer. Indeed, the future trials FOxTROT 2 and 3 may include monitoring of ctDNA during NAC. Although preoperative treatment is not a good setting to assess MRD per se, it is possible that the efficacy of NAC could be monitored and guided by the dynamics of ctDNA levels.
As mentioned previously, microsatellite instability has emerged as a prognostic and possible predictive marker in early stage colon cancer and should be considered part of the treatment algorithm in these patients. Patients with stage II and low risk stage III, MSI-High (MSI-H) colon cancer appear to have a lower risk of recurrence than cases with MSS tumors. Results from several studies have also suggested a lack of benefit of ACT with single fluorouracil (5-FU) in patients with MSI-H cancers [Citation8–12].
Also, therapies targeting Her-2, NTRAK fusion, KRAS G12C or BRAF-mutation may be included in future personalized treatment algorithms for subgroups of patients with suitable tumor characteristics for these treatments, as there are studies that suggest efficacy in the palliative setting [Citation49–51].
Concerning follow-up and prediction of survival, longitudinal ctDNA measurements seem to outperform CEA in RFS prediction according to early results that were presented at GI ASCO 2021 [Citation52]. However, results from additional trials need to prove the relation of clearance of ctDNA during ACT and DFS/OS.
Conclusion
Analysis of ctDNA shows great promise as a marker for both prognosis and response to treatment in colon cancer. Future studies will show whether we can use this technique to tailor treatment for patients in the adjuvant and neoadjuvant settings and whether this will translate into improved survival. Although ctDNA presently has some limitations, it has the potential to revolutionize treatment for patients with early stage colon cancer.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- (RCC) RCis. Cancer i Sverige Registerdata över förekomst och dödlighet 1970-2017. 2020.
- Edwards BK, Ward E, Kohler BA, et al. Annual report to the nation on the status of cancer, 1975-2006, featuring colorectal cancer trends and impact of interventions (risk factors, screening, and treatment) to reduce future rates. Cancer. 2010;116(3):544–573.
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424.
- Seymour MT, Morton D, obotIFT I. FOxTROT: an international randomised controlled trial in 1052 patients (pts) evaluating neoadjuvant chemotherapy (NAC) for Colon cancer. J Clin Oncol. 2019;37(15_suppl):3504–3504.
- Amin MB, Greene FL, Edge SB, et al. The eighth edition AJCC cancer staging manual: continuing to build a bridge from a population-based to a more "personalized" approach to cancer staging. CA Cancer J Clin. 2017;67(2):93–99.
- Schmoll HJ, Van Cutsem E, Stein A, et al. ESMO consensus guidelines for management of patients with colon and rectal cancer. a personalized approach to clinical decision making. Ann Oncol. 2012;23(10):2479–2516.
- Nicholson BD, Shinkins B, Pathiraja I, et al. Blood CEA levels for detecting recurrent colorectal cancer. Cochrane Database Syst Rev. 2015;2015(12):Cd011134.
- Popat S, Hubner R, Houlston RS. Systematic review of microsatellite instability and colorectal cancer prognosis. J Clin Oncol. 2005;23(3):609–618.
- Jover R, Zapater P, Castells A, et al. Mismatch repair status in the prediction of benefit from adjuvant fluorouracil chemotherapy in colorectal cancer. Gut. 2006;55(6):848–855.
- Sargent DJ, Marsoni S, Monges G, et al. Defective mismatch repair as a predictive marker for lack of efficacy of fluorouracil-based adjuvant therapy in Colon cancer. J Clin Oncol. 2010;28(20):3219–3226.
- Ribic CM, Sargent DJ, Moore MJ, et al. Tumor microsatellite-instability status as a predictor of benefit from fluorouracil-based adjuvant chemotherapy for Colon cancer. N Engl J Med. 2003;349(3):247–257.
- Cohen R, Taieb J, Fiskum J, et al. Microsatellite instability in patients with stage III Colon cancer receiving fluoropyrimidine with or without oxaliplatin: an ACCENT pooled analysis of 12 adjuvant trials. J Clin Oncol. 2021;39(6):642–651.
- Mlecnik B, Bifulco C, Bindea G, et al. Multicenter international society for immunotherapy of cancer study of the consensus immunoscore for the prediction of survival and response to chemotherapy in stage III Colon cancer. J Clin Oncol. 2020;38(31):3638–3651.
- Leon SA, Shapiro B, Sklaroff DM, et al. Free DNA in the serum of cancer patients and the effect of therapy. Cancer Res. 1977;37(3):646–650.
- Stroun M, Anker P, Lyautey J, et al. Isolation and characterization of DNA from the plasma of cancer patients. Eur J Cancer Clin Oncol. 1987;23(6):707–712.
- Corcoran RB, Chabner BA. Application of cell-free DNA analysis to cancer treatment. N Engl J Med. 2018;379(18):1754–1765.
- Reinert T, Henriksen TV, Christensen E, et al. Analysis of plasma cell-free DNA by ultradeep sequencing in patients with stages I to III colorectal cancer. JAMA Oncol. 2019;5(8):1124–1131.
- Tarazona N, Gimeno-Valiente F, Gambardella V, et al. Targeted next-generation sequencing of circulating-tumor DNA for tracking minimal residual disease in localized Colon cancer. Ann Oncol. 2019;30(11):1804–1812.
- Tie J, Cohen JD, Wang Y, et al. Circulating tumor DNA analyses as markers of recurrence risk and benefit of adjuvant therapy for stage III Colon cancer. JAMA Oncol. 2019;5(12):1710.
- Tie J, Wang Y, Tomasetti C, et al. Circulating tumor DNA analysis detects minimal residual disease and predicts recurrence in patients with stage II Colon cancer. Sci Transl Med. 2016;8(346):346ra92.
- Wang Y, Li L, Cohen JD, et al. Prognostic potential of circulating tumor DNA measurement in postoperative surveillance of nonmetastatic colorectal cancer. JAMA Oncol. 2019;5(8):1118.
- Taieb J, Taly V, Vernerey D, et al. Analysis of circulating tumour DNA (ctDNA) from patients enrolled in the IDEA-FRANCE phase III trial: prognostic and predictive value for adjuvant treatment duration. Ann Oncol. 2019;30:v867.
- Circulating tumour DNA based decision for adjuvant treatment in colon cancer stage II Evaluation. https://ClinicalTrials.gov/show/NCT04089631.
- Circulating tumor DNA based decision for adjuvant treatment in colon cancer stage II. https://ClinicalTrials.gov/show/NCT04120701.
- Taïeb J, Benhaim L, Laurent Puig P, et al. Decision for adjuvant treatment in stage II colon cancer based on circulating tumor DNA: the CIRCULATE-PRODIGE 70 trial. Dig Liver Dis. 2020;52(7):730–733.
- Tracking mutations in cell free tumour DNA to predict relapse in early colorectal cancer. https://ClinicalTrials.gov/show/NCT04050345.
- Anandappa G, Starling N, Peckitt C, et al. TRACC: tracking mutations in cell-free DNA to predict relapse in early colorectal cancer—a randomized study of circulating tumour DNA (ctDNA) guided adjuvant chemotherapy versus standard of care chemotherapy after curative surgery in patients with high risk stage II or stage III colorectal cancer (CRC). J Clin Oncol. 2020;38(15_suppl):TPS4120–TPS.
- IMPROVE intervention trial implementing non-invasive circulating tumor DNA analysis to optimize the operative and postoperative treatment for patients with colorectal cancer. https://ClinicalTrials.gov/show/NCT03748680.
- Nors J, Henriksen TV, Gotschalck KA, et al. IMPROVE-IT2: implementing noninvasive circulating tumor DNA analysis to optimize the operative and postoperative treatment for patients with colorectal cancer - intervention trial 2. Study protocol. Acta Oncol. 2020;59(3):336–341.
- Post-surgical liquid biopsy-guided treatment of stage III and high-risk stage II colon cancer patients: the PEGASUS trial. https://ClinicalTrials.gov/show/NCT04259944.
- Schraa SJ, van Rooijen KL, van der Kruijssen DEW, et al. Circulating tumor DNA guided adjuvant chemotherapy in stage II Colon cancer (MEDOCC-CrEATE): study protocol for a trial within a cohort study. BMC Cancer. 2020;20(1):790.
- Circulating Tumour DNA (ctDNA) as a prognostic and predictive marker in colorectal cancer – a pilot study. https://ClinicalTrials.gov/show/NCT04726800.
- Parsons HA, Rhoades J, Reed SC, et al. Sensitive detection of minimal residual disease in patients treated for Early-Stage breast cancer. Clin Cancer Res. 2020;26(11):2556–2564.
- Australian New Zealand Clinical Trials Registry [Internet]: Sydney (NSW): NHMRC Clinical Trials Centre UoSA-. Circulating tumour DNA (ctDNA) analysis informing adjuvant chemotherapy in Stage II Colon Cancer. 2015 [updated 2015 Mar 13]. https://www.anzctr.org.au/Trial/Registration/TrialReview.aspx?ACTRN=12615000381583
- Australian New Zealand Clinical Trials Registry [Internet]: Sydney (NSW): NHMRC Clinical Trials Centre UoSA-. Circulating Tumour DNA Analysis Informing Adjuvant Chemotherapy in Stage III Colon Cancer: A Multicentre Phase II/III Randomised Controlled Study (DYNAMIC-III): https://www.anzctr.org.au/Trial/Registration/TrialReview.aspx?id=373948. 2017. [updated 2017 Nov 8].
- Australian New Zealand Clinical Trials Registry [Internet]: Sydney (NSW): NHMRC Clinical Trials Centre UoSA-. Use of circulating tumour DNA (ctDNA) results to inform the decision for adjuvant chemotherapy in patients with locally advanced rectal cancer who have been treated with pre-operative chemo-radiation and surgery. http://www.anzctr.org.au/Trial/Registration/TrialReview.aspx?ACTRN=12617001560381. 2017. [updated 2017 Aug 19].
- Circulating tumor DNA testing in predicting treatment for patients with stage IIA colon cancer after surgery. https://ClinicalTrials.gov/show/NCT04068103.
- Dasari A. Circulating tumor DNA Alexandria: gastrointestinal cancers symposium. 2021. [cited 2021 Feb 16]. Available from: https://meetinglibrary.asco.org/record/193213/video.
- Taniguchi H, Nakamura Y, Kotani D, et al. CIRCULATE-Japan: circulating tumor DNA-guided adaptive platform trials to refine adjuvant therapy for colorectal cancer. Cancer Sci. 2021;112(7):2915–2920.
- Overman MJ. Discussion - oral abstract session: anal and colorectal cancer Alexandria: gastrointestinal cancers symposium. 2021. [cited 2021 Feb 10]. Available from: https://meetinglibrary.asco.org/record/194481/video.
- Phallen J, Sausen M, Adleff V, et al. Direct detection of early-stage cancers using circulating tumor DNA. Sci Transl Med. 2017;9(403):eaan2415.
- Schmitt MW, Kennedy SR, Salk JJ, et al. Detection of ultra-rare mutations by next-generation sequencing. Proc Natl Acad Sci USA. 2012;109(36):14508–14513.
- Diehl F, Schmidt K, Choti MA, et al. Circulating mutant DNA to assess tumor dynamics. Nat Med. 2008;14(9):985–990.
- Diehl F, Li M, Dressman D, et al. Detection and quantification of mutations in the plasma of patients with colorectal tumors. Proc Natl Acad Sci USA. 2005;102(45):16368–16373.
- Denis JA, Guillerm E, Coulet F, et al. The role of BEAMing and digital PCR for multiplexed analysis in molecular oncology in the era of Next-Generation sequencing. Mol Diagn Ther. 2017;21(6):587–600.
- Snyder MW, Kircher M, Hill AJ, et al. Cell-free DNA comprises an in vivo nucleosome footprint that informs its tissues-of-origin. Cell. 2016;164(1–2):57–68.
- Liu MC, Oxnard GR, Klein EA, et al. Sensitive and specific multi-cancer detection and localization using methylation signatures in cell-free DNA. Ann Oncol. 2020;31(6):745–759.
- Shen SY, Singhania R, Fehringer G, et al. Sensitive tumour detection and classification using plasma cell-free DNA methylomes. Nature. 2018;563(7732):579–583.
- Sartore-Bianchi A, Trusolino L, Martino C, et al. Dual-targeted therapy with trastuzumab and lapatinib in treatment-refractory, KRAS codon 12/13 wild-type, HER2-positive metastatic colorectal cancer (HERACLES): a proof-of-concept, multicentre, open-label, phase 2 trial. Lancet Oncol. 2016;17(6):738–746.
- Meric-Bernstam F, Hurwitz H, Raghav KPS, et al. Pertuzumab plus trastuzumab for HER2-amplified metastatic colorectal cancer (MyPathway): an updated report from a multicentre, open-label, phase 2a, multiple basket study. Lancet Oncol. 2019;20(4):518–530.
- Van Cutsem E, Huijberts S, Grothey A, et al. Binimetinib, encorafenib, and cetuximab triplet therapy for patients with BRAF V600E-Mutant metastatic colorectal cancer: safety lead-in results from the phase III BEACON colorectal cancer study. J Clin Oncol. 2019;37(17):1460–1469.
- Henriksen TV, Tarazona N, Reinert T, et al. Circulating tumor DNA analysis for assessment of recurrence risk, benefit of adjuvant therapy, and early relapse detection after treatment in colorectal cancer patients. J Clin Oncol. 2021;39(3_suppl):11–11.
