Abstract
Background
Detectable circulating tumor DNA (ctDNA) has been associated with worse prognosis in melanoma patients.
Material and methods
We studied plasma ctDNA as a prognostic biomarker in 19 patients with metastatic melanoma and a detectable tumor mutation (13 BRAF, 5 NRAS, and 1 KRAS). Patients had received chemotherapy, interferon-alpha, and vemurafenib in a prospective clinical trial. Mutant allele frequency (MAF %) was determined with droplet digital PCR from pretreatment and sequential plasma samples.
Results
Higher pretreatment plasma ctDNA levels (MAF ≥3%) and detectable plasma ctDNA levels (MAF >0%) at the time of radiologically confirmed best objective response were associated with poor prognosis even when accounting for other relevant prognostic factors including performance status, tumor mutation, metastasis stage, and lactate dehydrogenase levels in multivariable analysis.
Conclusion
Higher pretreatment plasma ctDNA levels and sustained detectable plasma ctDNA levels during treatment indicated poor prognosis in metastatic melanoma patients.
Background
Circulating tumor DNA (ctDNA) is an emerging prognostic biomarker for multiple cancer types [Citation1,Citation2]. Cancer cells release DNA fragments into the bloodstream and this cell-free ctDNA containing tumor-specific mutations can be detected using polymerase chain reaction (PCR) or next-generation DNA sequencing (NGS) [Citation2,Citation3]. Besides providing prognostic information, ctDNA can reveal therapeutically targetable tumor mutations and may reinforce radiological evaluation of tumor response in several cancers such as melanoma, NSCLC, colorectal, and breast cancer [Citation3–6].
More than two-thirds of cutaneous melanomas harbor a detectable tumor mutation, usually in BRAF (35–60%) or NRAS (15–28%) genes, that can be potentially monitored by liquid biopsies [Citation7–9]. Generally, plasma ctDNA reflects tumor volume and metabolic activity although some tumors shed only low amounts of ctDNA [Citation2]. PCR-based methods have revealed detectable ctDNA from plasma samples in 37.5−93% of stage IV [Citation10–14] and 11 − 37% of stage II and III melanoma patients with a known tumor mutation [Citation15,Citation16]. A positive correlation with detectable plasma ctDNA and higher tumor stage [Citation11,Citation17,Citation18], tumor burden (metabolic tumor volume assessed by 18FDG-PET-CT, sum of target lesions, and number of metastatic sites [Citation10–14,Citation18–20]), the presence of visceral metastases [Citation11,Citation14], and elevated LDH [Citation11,Citation20] has been observed in metastatic melanoma patients.
Immune checkpoint inhibitors (ICI) as well as BRAF and MEK inhibitors have replaced chemotherapy in the treatment of metastatic melanoma. Detectable plasma ctDNA has predicted worse prognosis in stage IV melanomas treated with BRAF and MEK inhibitors [Citation10,Citation11,Citation13], ICI [Citation12,Citation20], and bevacizumab [Citation21] as well as shorter disease-free survival and OS in stage II and III melanomas indicating “molecular residual disease” after complete surgical resection [Citation15,Citation16]. Increasing ctDNA levels during treatment indicated worse prognosis and ctDNA levels paralleled radiological tumor responses in metastatic melanomas treated with ICI [Citation12,Citation22]. Detectable pretreatment plasma ctDNA predicted disease progression (PD) and increasing ctDNA levels preceded radiologically confirmed PD in advanced melanoma patients treated with targeted therapies and ICI [Citation18].
In this study, we aimed to evaluate plasma ctDNA as a prognostic biomarker in metastatic melanoma patients treated with chemotherapy, interferon-alpha, and BRAF inhibitor vemurafenib in a prospective clinical trial [Citation23].
Material and methods
Study design
The COBRA trial was a nationwide prospective study enrolling 38 previously untreated, metastatic cutaneous melanoma patients in Finland between February 2014 and March 2016. Patients with asymptomatic brain metastases were eligible. The end of follow-up was in September 2019. All patients received chemotherapy (temozolomide, vincristine, and lomustine) with interferon-alpha (TOL-IFN) every four weeks for the maximum of six cycles followed by IFN maintenance therapy. Vemurafenib 960 mg twice a day was added for patients with BRAFv600 mutated melanomas after two cycles of TOL-IFN. The trial design, treatment, efficacy, and safety results were reported previously [Citation23].
BRAF mutations were analyzed at screening period from tumor tissue specimen using the fully integrated, real-time PCR-based IdyllaTM system (Biocartis, Belgium) or a cancer-targeted NGS panel (Ion Ampliseq Cancer Hotspot Panel v2, ThermoFisher Scientific, USA). Non-BRAF mutations were analyzed from tumor tissue specimen using the same NGS panel at Helsinki University Hospital. Pretreatment plasma samples, as well as sequential ones during study treatment, were collected in connection to radiological response evaluations. The centralized analysis of plasma samples was retrospectively performed at Helsinki University Hospital. Droplet digital PCR platform was used to quantify tumor-specific mutant allele frequency (MAF%) from pretreatment and sequential plasma samples of the patients with a known tumor mutation. Pretreatment plasma samples of the patients without a known tumor mutation were analyzed with NGS (Ion AmpliSeq Cancer Hotspot Panel v2) to reveal potentially undetected tumor mutations. Blood sample preparation, NGS, and ddPCR analyses were performed as described previously [Citation24]. Targeted mutation probes for BRAF, NRAS, IDH-1, and KRAS mutations were designed and prevalidated by Bio-Rad (Bio-Rad Laboratories, USA).
Statistical methods
The results of continuous variables are presented as median (range) and those of categorical variables as numbers and percentages. OS was measured from day 1 of TOL-IFN treatment to the date of death or the last follow-up visit. PFS was calculated similarly to the date of disease progression or the last follow-up visit. Kaplan-Meier curves were used to illustrate univariate analyses of PFS and OS. Kaplan-Meier estimates of OS and PFS are presented with 95% confidence intervals (95% CI) and the log-rank test was used to calculate statistical significance. Pearson’s two-sided Chi-Square test was used to calculate statistical differences in categorical variables and the one-way analysis of variance (ANOVA) was used for continuous variables. Cox regression analysis was performed to analyze the association of clinical characteristics with PFS and OS. All analyses were carried out with the IBM SPSS Statistics Version 27.
Ethical approval
The COBRA trial was approved by the Institutional Review Board and Ethics Committee of Helsinki University Hospital and registered to the European Union Drug Regulating Authorities Clinical Trials Database (Eudra CT study number 2013-000280-84). The trial was conducted in accordance with the Declaration of Helsinki and Good Clinical Practice Guidelines. All patients provided written informed consent. Data was anonymized before statistical analyses and handled in a manner that met general data protection regulations.
Results
Patient characteristics
The COBRA trial enrolled 38 patients of whom 23 (61%) had a detectable tumor mutation. Four patients were excluded due to lacking pretreatment and sequential plasma samples. Thus, 74 plasma samples from 19 patients with a known tumor mutation (11 BRAFV600E, 2 BRAFV600K, 2NRASQ61K, 3 NRASQ61R, and 1 KRASA146V) were analyzed with ddPCR to quantify tumor-specific ctDNA levels (MAF%) during study treatment. The median number of samples per patient was 3 (range 1 − 10).
Clinical characteristics of 19 patients included in the ctDNA analysis are described in Supplementary Table 1. Pretreatment MAF values ranged from 0% to 62% (median MAF 3.3%). 15 patients (79%) had detectable pretreatment plasma ctDNA (MAF >0%) levels. Detectable pretreatment plasma ctDNA (MAF >0% vs 0%) was associated with diminished ECOG performance status (0 vs ≥1, p < 0.001) but not with age, sex, LDH level, M-stage, tumor mutation (BRAF/non-BRAF), subsequent cancer therapy and subsequent ICI therapy. When MAF% cutoff was set to the observed median MAF, there were no significant associations with ctDNA levels (MAF ≥3% vs MAF <3%) and baseline clinical characteristics or subsequent cancer therapy.
Best objective response according to pretreatment plasma ctDNA levels
Among all patients in the ctDNA analysis, objective response rate (ORR) was 37%, disease control rate (DCR) 47%, and median time to best objective response (BOR) 1.9 (0.7 − 5.3) months. Patients with undetectable pretreatment ctDNA levels did not have significantly better objective response rate (ORR) compared to patients with detectable pretreatment ctDNA (50% vs 33%, p = 0.54) but had better disease control rate (DCR) (100% vs 33%, p = 0.018). Similarly, patients with pretreatment MAF <3% did not have better ORR (56% vs 20%, p = 0.11) but had better DCR (78% vs 20%, p = 0.012) compared to patients with MAF ≥3%. In BRAF-mutated melanomas, ORR was 54%, DCR 62%, and median time to BOR 3.5 (0.7 − 5.3) months. Undetectable pretreatment ctDNA was not predictive for better ORR (67% vs 50%, p = 0.61) nor DCR (100% vs 50%, p = 0.12) in BRAF mutated melanomas. However, patients with pretreatment BRAF MAF <3% had better ORR (83% vs 29%, p = 0.048) and better DCR (100% vs 29%, p = 0.008) compared to patients with pretreatment BRAF MAF ≥3%.
PFS and OS according to plasma ctDNA levels
At the end of follow-up, three patients with BRAFV600E mutated melanomas were alive and progression-free. The median PFS was 2.1 (0.7 − 64.5) months for all patients and 5.6 (0.7 − 64.5) months for BRAF-mutated melanomas. The median OS was 12.9 (0.7 − 64.5) months for all patients and 13.1 (0.7 − 64.5) months for BRAF-mutated melanomas.
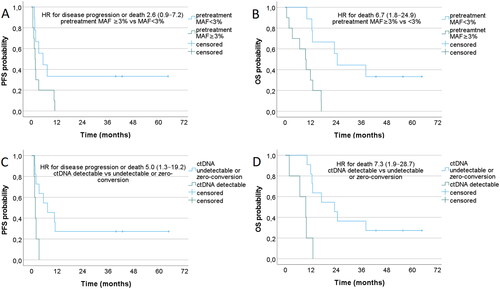
Higher pretreatment ctDNA levels (MAF ≥3% vs MAF <3%) were associated with inferior OS among all 19 ctDNA patients () and among 13 BRAF mutated melanomas (Supplementary Figure 1(B)). Higher pretreatment ctDNA levels were not significantly associated with inferior PFS ( and Supplementary Figure 1(A)). Detectable ctDNA levels (MAF >0% vs 0%) at the time of BOR were associated with inferior PFS as well as inferior OS among all 19 ctDNA patients () although this finding did not reach statistical significance among 13 BRAF mutated melanomas (Supplementary Figure 1(C,D)). PFS and OS according to detectable vs undetectable pretreatment ctDNA levels (MAF >0% vs 0%) are shown in Supplementary Figure 2.
Multivariable cox regression models for PFS and OS
In multivariable Cox regression analysis, higher levels of pretreatment ctDNA (MAF ≥3% vs <3%) and detectable ctDNA at BOR (detectable vs undetectable/zero-conversion) were independently associated with inferior PFS and OS when adjusted for other relevant clinical characteristics including age, sex, tumor mutation (BRAF/nonBRAF), LDH level, M-stage, ECOG performance status, and subsequent therapies (). Detectable pretreatment ctDNA (MAF >0% vs 0%) was not independently associated with PFS or OS in the multivariable analysis.
Table 1. Multivariable Cox regression models for progression-free survival and overall survival.
Discussion
BRAF and MEK inhibitors and ICI have remarkably improved survival in metastatic melanoma even in patients with poor prognostic features such as elevated LDH and CRP, more than three metastatic sites, visceral metastases, and brain metastases [Citation25–28]. In addition to traditional risk factors, detectable plasma ctDNA is associated with worse prognosis in advanced cutaneous melanomas treated with BRAF plus MEK inhibitors [Citation10,Citation11,Citation13] and ICI [Citation12,Citation20] as well as in uveal melanomas treated with HDAC inhibitor (entinostat) and pembrolizumab [Citation29].
In this study, we discovered, that higher pretreatment ctDNA levels (MAF ≥3%) and sustained detectable ctDNA (MAF >0%) at the time of BOR were associated with worse prognosis in metastatic cutaneous melanoma patients treated with chemoimmunotherapy ± vemurafenib. One may argue that ctDNA does not add prognostic value to well-established risk factors, such as tumor burden and LDH level. However, we found that higher pretreatment ctDNA levels (MAF ≥3%) and sustained detectable ctDNA at BOR were independently associated with inferior PFS and OS after adjusted for other relevant risk factors including ECOG performance status, LDH, and M-stage. Similarly, zero-conversion of detectable pretreatment plasma ctDNA during treatment has indicated better prognosis for metastatic melanoma patients treated with PD-1 inhibitors or BRAF + MEK inhibitors [Citation13,Citation20]. Interestingly, zero-conversion was associated with prolonged PFS and OS in patients with elevated LDH but not in patients with normal LDH [Citation13]. Thus, ctDNA seems to provide clinically relevant information in addition to traditional risk factors.
The limitations of our study include small number of patients enrolled in the COBRA trial and limited number of plasma samples available for ddPCR analysis. This prospective trial was initiated in 2014 when BRAF and MEK inhibitors were not available outside clinical trials in Finland. Patients with BRAFV600 mutated melanomas received vemurafenib within the trial but BRAF-negative patients lacked modern treatment options (ICI) at that time. Subsequent cancer therapies included modern treatments which could have prolonged the OS of those patients. Despite an old-fashioned study treatment, the results of ctDNA analyses in this study were in concordance with other studies establishing ctDNA as a prognostic biomarker in metastatic melanoma regardless of the choice of systemic treatment.
In conclusion, higher pretreatment ctDNA levels (MAF ≥3%) and sustained detectable ctDNA at BOR were poor prognostic factors in metastatic melanoma patients who had received chemoimmunotherapy ± BRAF inhibitor. Metastatic melanoma patients with persistently detectable ctDNA levels during systemic treatment may need more frequent radiological response evaluation to confirm disease progression and perhaps other treatment options.
Author contributions
KEM had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis. MH, SM, and PV: study concept and design. KEM, SM, SK, EA, PV, SR, TS, LT, MSV, KTK, JK, LK, and MH: acquisition of data. KEM, SM, PV, and MH: analysis and interpretation of data. KEM, SM, SK, EA, PV, SR, TS, LT, MSV, KTK, JK, LK, KA, and MH: drafting of the manuscript. KEM: statistical analysis. MH, PV, SK, KA, and KEM: Obtaining funding. MH, PV, and SK: supervision.
Supplemental Material
Download MS Word (12.5 KB)Supplemental Material
Download TIFF Image (333.1 KB)Supplemental Material
Download TIFF Image (346 KB)Disclosure statement
F. Hoffmann-La Roche Ltd provided vemurafenib for study patients until reimbursement in Finland in 2016 and the Finnish Melanoma Group received a grant from F. Hoffmann-La Roche Ltd. KEM reports grants from the Finnish Melanoma Group and the Cancer Society of South-West Finland and has received consulting or advisory honoraria from Astellas, Bayer, Bristol-Myers Squibb, Ipsen, Jansen, Merck Sharp & Dohme, Merck − Pfizer alliance, Novartis, Roche, and Sanofi. SM has received consulting or advisory honoraria from Amgen, Bristol-Myers Squibb, Merck Sharp & Dohme, Merck Group, Novartis, Roche and Sanofi; Speakers’ Bureau honoraria from Bristol – Myers Squibb, Merck Sharp & Dohme, and Sanofi. PV has received consulting or advisory honoraria from Merck, Bristol-Myers Squibb, Ipsen, Novartis, and Roche; speakers’ bureau honoraria from Merck, Roche, and Bristol-Myers Squibb. MSV reports financial support for scientific meetings from Novartis and Merck-Pfizer and advisory honoraria from Novartis. JK reports grants and personal fees from Roche, BMS, Merck, and Novartis, personal fees all outside the submitted work. MH has received consulting or advisory honoraria from Merck, Bristol-Myers Squibb, Incyte, Varian, Novartis, and Roche; speakers’ bureau honoraria from Merck, Novartis, and Bristol-Myers Squibb. SK, EA, SR, TS, LT, KTK, LK, and KA have no conflicts of interest to declare.
Data availability statement
Data are available upon reasonable request to corresponding author.
References
- Bettegowda C, Sausen M, Leary RJ, et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med. 2014;6(224):224ra24.
- Keller L, Belloum Y, Wikman H, et al. Clinical relevance of blood-based ctDNA analysis: mutation detection and beyond. Br J Cancer. 2021;124(2):345–358.
- Kilgour E, Rothwell DG, Brady G, et al. Liquid Biopsy-Based biomarkers of treatment response and resistance. Cancer Cell. 2020;37(4):485–495.
- Gray ES, Rizos H, Reid AL, et al. Circulating tumor DNA to monitor treatment response and detect acquired resistance in patients with metastatic melanoma. Oncotarget. 2015;6(39):42008–42018.
- Dasari A, Morris VK, Allegra CJ, et al. ctDNA applications and integration in colorectal cancer: an NCI Colon and rectal–anal task forces whitepaper. Nat Rev Clin Oncol. 2020;17(12):757–770.
- Dawson S-J, Tsui DWY, Murtaza M, et al. Analysis of circulating tumor DNA to monitor metastatic breast cancer. N Engl J Med. 2013;368(13):1199–1209.
- Cancer genome atlas network. Genomic classification of cutaneous melanoma. Cell. 2015;161(7):1681–1696.
- Ny L, Hernberg M, Nyakas M, et al. BRAF mutational status as a prognostic marker for survival in malignant melanoma: a systematic review and meta-analysis. Acta Oncol. 2020;59(7):833–844.
- Lee J-H, Choi J-W, Kim Y-S. Frequencies of BRAF and NRAS mutations are different in histological types and sites of origin of cutaneous melanoma: a meta-analysis. Br J Dermatol. 2011;164(4):776–784.
- Ascierto PA, Minor D, Ribas A, et al. Phase II trial (BREAK-2) of the BRAF inhibitor dabrafenib (GSK2118436) in patients with metastatic melanoma. J Clin Oncol. 2013;31(26):3205–3211.
- Santiago-Walker A, Gagnon R, Mazumdar J, et al. Correlation of BRAF mutation status in Circulating-Free DNA and tumor and association with clinical outcome across four BRAFi and MEKi clinical trials. Clin Cancer Res. 2016;22(3):567–574.
- Seremet T, Jansen Y, Planken S, et al. Undetectable circulating tumor DNA (ctDNA) levels correlate with favorable outcome in metastatic melanoma patients treated with anti-PD1 therapy. J Transl Med. 2019;17(1):303.
- Syeda MM, Wiggins JM, Corless BC, et al. Circulating tumour DNA in patients with advanced melanoma treated with dabrafenib or dabrafenib plus trametinib: a clinical validation study. Lancet Oncol. 2021;22(3):370–380.
- Rowe SP, Luber B, Makell M, et al. From validity to clinical utility: the influence of circulating tumor DNA on melanoma patient management in a real-world setting. Mol Oncol. 2018;12(10):1661–1672.
- Lee RJ, Gremel G, Marshall A, et al. Circulating tumor DNA predicts survival in patients with resected high-risk stage II/III melanoma. Ann Oncol. 2018;29(2):490–496.
- Tan L, Sandhu S, Lee RJ, et al. Prediction and monitoring of relapse in stage III melanoma using circulating tumor DNA. Ann Oncol. 2019;30(5):804–814.
- Gonzalez-Cao M, Mayo-de-Las-Casas C, Molina-Vila MA, et al. BRAF mutation analysis in circulating free tumor DNA of melanoma patients treated with BRAF inhibitors. Melanoma Res. 2015;25(6):486–495.
- BRAF mutation analysis in circulating free tumor DNA of melanoma patients treated with BRAF inhibitors. Melanoma Res. 2015;25(6):486–495.
- Váraljai R, Wistuba-Hamprecht K, Seremet T, et al. Application of circulating Cell-Free tumor DNA profiles for therapeutic monitoring and outcome prediction in genetically heterogeneous metastatic melanoma. J Clin Oncol Precis Oncol. 2019;3(3):1–10.
- McEvoy AC, Warburton L, Al-Ogaili Z, et al. Correlation between circulating tumour DNA and metabolic tumour burden in metastatic melanoma patients. BMC Cancer. 2018;18(1):726.
- Lee JH, Long GV, Boyd S, et al. Circulating tumour DNA predicts response to anti-PD1 antibodies in metastatic melanoma. Ann Oncol. 2017;28(5):1130–1136.
- Forthun RB, Hovland R, Schuster C, et al. ctDNA detected by ddPCR reveals changes in tumour load in metastatic malignant melanoma treated with bevacizumab. Sci Rep. 2019;9(1):17471.
- Seremet T, Planken S, Schreuer M, et al. Illustrative cases for monitoring by quantitative analysis of BRAF/NRAS ctDNA mutations in liquid biopsies of metastatic melanoma patients who gained clinical benefits from anti-PD1 antibody therapy. Melanoma Res. 2018;28(1):65–70.
- Mattila KE, Vihinen P, Ramadan S, et al. Combination chemotherapy with temozolomide, lomustine, vincristine and interferon-alpha (TOL-IFN) plus vemurafenib or TOL-IFN as first-line treatment for patients with advanced melanoma. Acta Oncol. 2020;59(3):310–314.
- Holm M, Andersson E, Osterlund E, et al. Detection of KRAS mutations in liquid biopsies from metastatic colorectal cancer patients using droplet digital PCR, idylla, and next generation sequencing. PLoS One. 2020;15(11):e0239819.
- Long GV, Grob J-J, Nathan P, et al. Factors predictive of response, disease progression, and overall survival after dabrafenib and trametinib combination treatment: a pooled analysis of individual patient data from randomised trials. Lancet Oncol. 2016;17(12):1743–1754.
- Weide B, Martens A, Hassel JC, et al. Baseline biomarkers for outcome of melanoma patients treated with pembrolizumab. Clin Cancer Res. 2016;22(22):5487–5496.
- Iivanainen S, Ahvonen J, Knuuttila A, et al. Elevated CRP levels indicate poor progression-free and overall survival on cancer patients treated with PD-1 inhibitors. ESMO Open. 2019;4(4):e000531.
- Ny L, Jespersen H, Karlsson J, et al. The PEMDAC phase 2 study of pembrolizumab and entinostat in patients with metastatic uveal melanoma. Nat Commun. 2021;12(1):5155.