Introduction
Lymphomas are a group of heterogeneous neoplasms in which lymphoid cells undergo malignant transformation [Citation1]. In the updated WHO and ICC classifications [Citation1,Citation2], >80 different subtypes are recognized. Mature B-cell lymphoma subtypes make up the majority of the cases, including, e.g., diffuse large B-cell lymphoma (DLBCL), chronic lymphocytic leukemia (CLL), and follicular lymphoma (FL). The introduction of more comprehensive sequencing techniques for genomic characterization has increased awareness that lymphomas are biologically distinct, even within the same established subtype [Citation3,Citation4].
Although lymphomas represent a group of highly chemo-sensitive malignancies, some patients do not respond to established chemo-immunotherapy or experience relapse after initial response, with much-worsened prognosis [Citation5,Citation6]. New targeted or cellular therapies have recently been approved or are being tested in clinical trials in treatment-refractory or relapsed disease with highly encouraging results, including, e.g., genetically modified autologous T cells targeting CD19 (CAR-T therapy) [Citation7] and bi-specific antibodies [Citation8]. Bruton’s tyrosine kinase inhibitors (BTKi) have been successfully introduced in the treatment of some lymphoma subtypes (e.g., CLL) but show only modest response in DLBCL and FL, although response rates may be higher in certain molecular subtypes [Citation9–11].
Only a few genetic alterations are currently incorporated into clinical decision-making and selection of treatment in lymphoproliferative malignancies; TP53 aberrations in CLL and MCL [Citation12], IGHV gene mutational status in CLL [Citation13], and MYC/BCL2 rearrangements in high-grade B-cell lymphoma [Citation11]. While gene expression can be used to identify prognostic subgroups (e.g., germinal-center and activated B-cell (GCB/ABC DLBCL)), recent landmark studies have suggested new molecular subgroups in DLBCL and CLL based on comprehensive genomic sequencing with potential implications for targeted therapy [Citation11,Citation14–16], however, their clinical usefulness is still to be determined. In addition, analysis of cell-free DNA (cfDNA) has emerged as a promising tool to monitor treatment response and measurable residual disease (MRD) in a more sensitive way than with current radiology-based methods [Citation17–20].
In the present prospective cohort study of newly diagnosed lymphoma patients in Stockholm, Sweden (the BioLymph study), we aim to increase knowledge and implementation of precision diagnostic tools and personalized medicine approaches across mature B- and T-cell lymphomas in a population-based setting. In this first report, we evaluate the feasibility of patient inclusion and targeted next-generation sequencing (NGS) among the first 100 included patients.
Materials and methods
Study design and patient recruitment
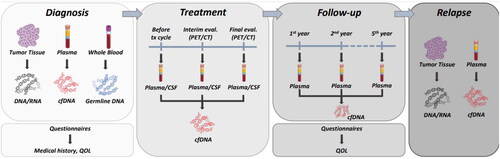
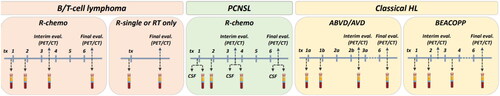
Adult patients (≥18 years) with newly diagnosed lymphoma at Karolinska University Hospital, Stockholm, during the study period are eligible for inclusion. Patients should have a representative tumor sample taken as part of routine diagnostic work-up to allow for the preparation of tumor DNA. Patients who do not understand the written Swedish language or with dementia or other CNS disease that could hamper provision of written informed consent are excluded. The inclusion period started 11 February 2019 and is planned to continue until December 2024 (aiming for the inclusion of 500 patients), with follow-up of five years. Patient participation entails preparation of DNA and RNA from the diagnostic tumor sample, donation of blood and plasma samples at diagnosis and during follow-up (), completion of questionnaires of medical history and quality-of-life (EORTC-QLQ30, fatigue, neuropathy) [Citation21,Citation22] and extraction of relevant information from medical records and national health care registers [Citation23]. At diagnosis, a whole blood sample is taken for analysis of germline DNA, and plasma samples are taken in EDTA tubes and stored for future proteomics analyses. Repeated plasma samples are taken for cfDNA analysis in blood collection tubes (STRECK, La Vista, USA) according to treatment-specific schedules (). During follow-up, plasma samples are repeated in case of progression/relapse or transformation. Annual disease-free follow-up includes plasma sampling and completion of quality-of-life questionnaires at 1, 2, and 5 years post-diagnosis.
Endpoints
Feasibility was evaluated by the proportion of patients that completed the first parts of the study protocol following inclusion and the proportion of patients with successful targeted NGS on tumor tissue. Endpoints further included the number, proportion and type of genomic driver alterations potentially relevant for diagnosis, prognosis and treatment prediction. Detailed specific aims and endpoints for the full study are listed in Supplementary Table 1. DNA is extracted from tumor tissue and whole blood (Qiasymphony, Qiagen). During the first year of patient inclusion, paired tumor and germline DNA samples were sequenced with the amplicon-based TruSight lymphoma gene panel (Illumina, San Diego, CA, USA) including 43 genes known to be recurrently mutated in lymphoma (Supplementary Table 2) [Citation24]. In the second year, we switched to a broader capture-based gene panel including 252 genes (GMS Lymphoid panel, Twist BioScience) (Supplementary Table 3) [Citation25]. [Citation26]. For validation purposes, a few samples (n = 6) were analyzed both with the TruSight and GMS-Lymphoid panels. In a second validation step, we also compared the NGS results of the GMS-Lymphoid panel on FF and FFPE tumor material from the same patients.
Bioinformatics and variant interpretation
Sequencing data were analyzed using standardized bioinformatic pipelines developed and maintained by SciLifeLab and the nf-core community [Citation27,Citation28]. Following further quality checks in the Scout program (available at https://github.com/Clinical-Genomics/scout), variant interpretation was carried out by molecular/clinical geneticists based on the 43 genes covered by the TruSight panel (Supplementary Table 2). Mutations were classified as clinically relevant if they were found to be pathogenic or likely pathogenic (class 4 or 5) according to adapted ACMG criteria [Citation29], in combination with a variant allele frequency (VAF) of ≥5%. Flagged variants were discussed at multidisciplinary molecular tumor boards (MTBs).
The study has been approved by the regional ethics board in Stockholm (ref no 2017/2538-31, approval date 19/02/2018). All participating patients provide written informed consent at inclusion. The study has been registered as ISRCTN12948913.
Results
Among the first 100 included patients, the majority were male (62%) and median age at diagnosis was 69 years (range 19–94) (). The most common lymphoma subtypes were DLBCL (33%), FL (19%) and Hodgkin lymphoma (HL, 18%). Almost all patients (≥96%) had peripheral blood samples taken at diagnosis in line with the study protocol. Over 90% of the patients also returned baseline medical history and quality-of-life questionnaires. Genomic DNA could be prepared from tumor tissue from 84% of the patients with B/T-cell lymphomas (n = 69/82) (HL cases, n = 18, were excluded due to low tumor cell content) (). DNA could not be prepared for some patients (n = 13) due to limited availability of material. In the initial year of the study, we performed the amplicon-based TruSight panel on DNA prepared from FF and FFPE tissue. Sequence data were successfully derived for 32 out of 33 patients. However, when analyzing DNA prepared from paired FFPE tissue samples the concordance with FF material was suboptimal with both false-positive and false-negative variants (exemplified in Supplementary Figure 1, right panel). Thus, we only considered results for patients with TruSight sequence results from FF tumor samples (n = 20).
Table 1. Overview of patient characteristics and materials collected at diagnosis, and targeted sequencing performed for the first 100 study participants in the BioLymph study.
Considering that FFPE tissue is available to a larger extent than FF tissue in lymphoma diagnostics, we next applied a hybrid-capture sequencing technology that is known to give a higher and more even sequencing yield also for FFPE samples (the GMS Lymphoid panel). In a subset of cases (n = 6), analysis with both panels across tissue specimens (trios of FF and FFPE tissue and blood) demonstrated a high concordance with similar sequencing results for the overlapping genes (exemplified in Supplementary Figure 1, left and middle panel). We further validated the results of the GMS Lymphoid panel in FF tumor tissue (without restriction to clinically relevant variants) in the same patient subset. The concordance of the NGS results using the GMS Lymphoid panel based on FF and FFPE tissue was 99.5% at VAF ≥5% (Supplementary Table 3, Supplementary Figure 2). In view of these reassuring results, we next analyzed FFPE tumor samples in 30 additional patients. Overall, using the GMS Lymphoid panel, 35/36 patients could be sequenced with average sequencing depth of ∼2000X and ∼99% of bases had 100-fold coverage. In total, considering both panels, clinically relevant mutations were found for 69% of patients with successful sequencing (38/55). Among all patients with at least one clinically relevant mutation, the median number of such mutations was 2.
Discussion
Implementation of precision medicine has already revolutionized care for some patients with hematological malignancies [Citation30]. For example, in several myeloid disorders, genetic aberrations help determine diagnosis, prognosis, and treatment for individual patients [Citation3,Citation30–33]. In contrast, for lymphoproliferative malignancies, only a few aberrations are currently used in clinical practice [Citation11–13]. However, it is evident that improved knowledge regarding the impact of genetic aberrations in lymphoma will make the classification and management of lymphomas more granular and personalized through improved prognostication and by enabling individualized treatment decisions [Citation3,Citation9,Citation16]. To ultimately achieve this, studies such as BioLymph are expected to be important.
As evident from this first report of 100 patients, the collection of samples and questionnaires followed the study protocol to a very high extent. Importantly, we also demonstrate the feasibility of NGS analyses from FFPE tumor material when using the hybrid-capture-based GMS-Lymphoid panel, for which analysis of DNA from FF and FFPE material provided highly concordant results. In other words, for most patients where FFPE tumor material is the only material available for diagnosis, NGS can still be performed. Among the patients with B/T-cell lymphoma in this first analysis, two-thirds (69%) had at least one clinically relevant genetic alteration in the tumor tissue. However, only 43 genes were evaluated (i.e., genes covered by both panels). In the next phase of the Biolymph study, we will expand the analysis to the full GMS Lymphoid panel and thus, we expect the number of cases with detected alterations, and the number of alterations per case, to increase. The GMS Lymphoid panel, here encompassing 252 genes, is being further developed to include other important features, such as structural aberrations (e.g., MYC, BCL2, and CCND1 translocations) and immunoglobulin and T-cell receptor gene rearrangements, all important biomarkers in lymphoma diagnostics.
An important challenge with the implementation of precision medicine in lymphoma is to define how specific molecular findings should be interpreted in the clinical routine. This is particularly relevant for cases with multiple genetic aberrations, where the presence of one aberration may affect the impact of another [Citation14]. To address this challenge, MTBs with multidisciplinary participation has proven helpful in the BioLymph study. Moreover, portals in which genomic data can be entered and processed through several online genetic databases leading to a report can be important future tools for clinical decision-making [Citation34]. Several studies have reported the potential of analysis of cfDNA to assess treatment response and aid diagnosis and early detection of relapse in lymphomas [Citation18,Citation35,Citation36]. For lymphomas that are anatomically hard to access, such as primary mediastinal B-cell lymphomas or CNS lymphomas, detection of tumor-derived cfDNA could also add diagnostic value. This may also be the case for classical HL where analyses of Hodgkin-Reed-Sternberg cells are complicated by their scarcity [Citation18,Citation37]. However, time points for when measurements of cfDNA are of the most value remain to be established, and more data and validation of analytical methods are needed to determine the clinical utility of this new tool. The use of questionnaires in BioLymph will enable studies on the association of lymphoma diagnosis and treatment with general well-being, fatigue, and neuropathy. In addition, the linkage of data from BioLymph to other national registers will offer opportunities to study an array of other adverse outcomes, both short and long-term.
In conclusion, with this first report, we show that the BioLymph study protocol and patient inclusion is feasible, that completion rates in the initial study phases are high, and that sequencing of tumor material adds clinically relevant information in at least two-thirds of the patients. In view of the integration within clinical health care and the confluence of multidisciplinary experts, the study promises to contribute substantially to the development and implementation of precision medicine approaches in lymphoma patient care in Sweden and abroad.
Supplemental Material
Download MS Word (20.1 KB)Supplemental Material
Download MS Word (283.5 KB)Acknowledgements
We are deeply indebted to the clinical research personnel at the departments of Hematology, Pathology, and Clinical Genetics for their work with patient inclusion and collection of materials in the present study, in particular Emilie Nelson, Anna-Lena Fridolfsson, Shaila Petersson, Lylie Mbuyi, Harriet Ryblom, and Hero Nikdin. We further acknowledge the excellent support by Clinical Genomics Stockholm at SciLifeLab in the NGS-related work of the study. Funding support is provided by Theme Cancer, Karolinska University Hospital (first 100 patients), Region Stockholm (No. 20190494, 500306 and FoUI-961732), King V Jubilee Fund (No. 214153), Medical Diagnostics Karolinska (No. 2022-3763), the Swedish Childhood Cancer Fund (No. TJ2021-0125, KP2021-0017) and through Genome Medicine Sweden (GMS). GMS is funded by the Swedish Innovation Agency Vinnova and through collaborating partners.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
Study data are available upon request if in line with ethical and legal permissions.
Correction Statement
This article has been corrected with minor changes. These changes do not impact the academic content of the article.
Additional information
Funding
References
- Alaggio R, Amador C, Anagnostopoulos I, et al. The 5th edition of the world health organization classification of haematolymphoid tumours: lymphoid neoplasms. Leukemia. 2022;36(7):1720–1748.
- Campo E, Jaffe ES, Cook JR, et al. The international consensus classification of mature lymphoid neoplasms: a report from the clinical advisory committee. Blood. 2022;140(11):1229–1253.
- de Leval L, Alizadeh AA, Bergsagel PL, et al. Genomic profiling for clinical decision making in lymphoid neoplasms. Blood. 2022;140(21):2193–2227.
- Mansouri L, Thorvaldsdottir B, Laidou S, et al. Precision diagnostics in lymphomas – Recent developments and future directions. Semin Cancer Biol. 2022;84:170–183.
- Harrysson S, Eloranta S, Ekberg S, et al. Incidence of relapsed/refractory diffuse large B-cell lymphoma (DLBCL) including CNS relapse in a population-based cohort of 4243 patients in Sweden. Blood Cancer J. 2021;11(1):9.
- Casulo C, Byrtek M, Dawson KL, et al. Early relapse of follicular lymphoma after rituximab plus cyclophosphamide, doxorubicin, vincristine, and prednisone defines patients at high risk for death: an analysis from the national lymphocare study. J Clin Oncol. 2015;33(23):2516–2522.
- Neelapu SS, Locke FL, Bartlett NL, et al. Axicabtagene ciloleucel CAR T-Cell therapy in refractory large B-cell lymphoma. N Engl J Med. 2017;377(26):2531–2544.
- Velasquez MP, Bonifant CL, Gottschalk S. Redirecting T cells to hematological malignancies with bispecific antibodies. Blood. 2018;131(1):30–38.
- Wilson WH, Wright GW, Huang DW, et al. Effect of ibrutinib with R-CHOP chemotherapy in genetic subtypes of DLBCL. Cancer Cell. 2021;39(12):1643–1653.e3.
- Byrd JC, Furman RR, Coutre SE, et al. Three-year follow-up of treatment-naïve and previously treated patients with CLL and SLL receiving single-agent ibrutinib. Blood. 2015;125(16):2497–2506.
- Sehn LH, Salles G. Diffuse large B-cell lymphoma. N Engl J Med. 2021;384(9):842–858.
- Campo E, Cymbalista F, Ghia P, et al. Aberrations in chronic lymphocytic leukemia: an overview of the clinical implications of improved diagnostics. Haematologica. 2018;103(12):1956–1968.
- Agathangelidis A, Chatzidimitriou A, Chatzikonstantinou T, et al. Immunoglobulin gene sequence analysis in chronic lymphocytic leukemia: the 2022 update of the recommendations by ERIC, the European research initiative on CLL. Leukemia. 2022;36(8):1961–1968.
- Wright GW, Huang DW, Phelan JD, et al. A probabilistic classification tool for genetic subtypes of diffuse large B cell lymphoma with therapeutic implications. Cancer Cell. 2020;37(4):551–568 e14.
- Knisbacher BA, Lin Z, Hahn CK, et al. Molecular map of chronic lymphocytic leukemia and its impact on outcome. Nat Genet. 2022;54(11):1664–1674.
- Chapuy B, Stewart C, Dunford AJ, et al. Molecular subtypes of diffuse large B cell lymphoma are associated with distinct pathogenic mechanisms and outcomes. Nat Med. 2018;24(5):679–690.
- Scherer F, Kurtz DM, Newman AM, et al. Distinct biological subtypes and patterns of genome evolution in lymphoma revealed by circulating tumor DNA. Sci Transl Med. 2016;8(364):364ra155.
- Spina V, Bruscaggin A, Cuccaro A, et al. Circulating tumor DNA reveals genetics, clonal evolution, and residual disease in classical Hodgkin lymphoma. Blood. 2018;131(22):2413–2425.
- Camus V, Viennot M, Lequesne J, et al. Targeted genotyping of circulating tumor DNA for classical Hodgkin lymphoma monitoring: a prospective study. Haematologica. 2021;106(1):154–162.
- Rivas-Delgado A, Nadeu F, Enjuanes A, et al. Mutational landscape and tumor burden assessed by cell-free DNA in diffuse large B-cell lymphoma in a population-based study. Clin Cancer Res. 2021;27(2):513–521.
- Sprangers MA, Cull A, Bjordal K, et al. The european organization for research and treatment of cancer. Approach to quality of life assessment: guidelines for developing questionnaire modules. EORTC study group on quality of life. Qual Life Res. 1993;2(4):287–295.
- Stefansson M, Nygren P. Oxaliplatin added to fluoropyrimidine for adjuvant treatment of colorectal cancer is associated with long-term impairment of peripheral nerve sensory function and quality of life. Acta Oncol. 2016;55(9–10):1227–1235.
- Ekberg S, E Smedby K, Glimelius I, et al. Trends in the prevalence, incidence and survival of non-Hodgkin lymphoma subtypes during the 21st century - a Swedish lymphoma register study. Br J Haematol. 2020;189(6):1083–1092.
- Rosenquist R, Rosenwald A, Du MQ, et al. Clinical impact of recurrently mutated genes on lymphoma diagnostics: state-of-the-art and beyond. Haematologica. 2016;101(9):1002–1009.
- Bonfiglio S, Sutton LA, Ljungström V, et al. BTK and PLCG2 remain unmutated in one third of patients with CLL relapsing on ibrutinib. Blood Adv. 2023. DOI:10.1182/bloodadvances.2022008821
- Horak P, Griffith M, Danos AM, et al. Standards for the classification of pathogenicity of somatic variants in cancer (oncogenicity): joint recommendations of clinical genome resource (ClinGen), cancer genomics consortium (CGC), and variant interpretation for cancer consortium (VICC). Genet Med. 2022;24(9):1991.
- Ewels PA, Peltzer A, Fillinger S, et al. The nf-core framework for community-curated bioinformatics pipelines. Nat Biotechnol. 2020;38(3):276–278.
- Foroughi-Asl H, Jeggari, Ashwini, Maqbool, Khurram, Ivanchuk, Vadym, Elhami, Keyvan, Wirta V. BALSAMIC: bioinformatic Analysis pipeLine for SomAtic MutatIons in Cancer (v10.0.3). 2022.
- Richards S, Aziz N, Bale S, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American college of medical genetics and genomics and the association for molecular pathology. Genet Med. 2015;17(5):405–424.
- Wasterlid T, Cavelier L, Haferlach C, et al. Application of precision medicine in clinical routine in haematology-challenges and opportunities. J Intern Med. 2022;292(2):243–261.
- Haferlach T. Advancing leukemia diagnostics: role of next generation sequencing (NGS) in acute myeloid leukemia. Hematol Rep. 2020;12(Suppl 1):8957.
- Hochhaus A, Masszi T, Giles FJ, et al. Treatment-free remission following frontline nilotinib in patients with chronic myeloid leukemia in chronic phase: results from the ENESTfreedom study. Leukemia. 2017;31(7):1525–1531.
- Duncavage EJ, Bagg A, Hasserjian RP, et al. Genomic profiling for clinical decision making in myeloid neoplasms and acute leukemia. Blood. 2022;140(21):2228–2247.
- Tamborero D, Dienstmann R, Rachid MH, et al. Support systems to guide clinical decision-making in precision oncology: the cancer core Europe molecular tumor board portal. Nat Med. 2020;26(7):992–994.
- Rossi D, Diop F, Spaccarotella E, et al. Diffuse large B-cell lymphoma genotyping on the liquid biopsy. Blood. 2017;129(14):1947–1957.
- Huet S, Salles G. Potential of circulating tumor DNA for the management of patients with lymphoma. J Clin Oncol Oncol Pract. 2020;16(9):561–568.
- Sarkozy C, Hung SS, Chavez EA, et al. Mutational landscape of gray zone lymphoma. Blood. 2021;137(13):1765–1776.