Abstract
Appropriate definitions of species limits are critical for scientific study and conservation management. Many subspecies of birds are defined on the basis of phenotypic variation. However, it has been suggested that following close examination many of these subspecies will be recognised as good species. North Island (NI) (Philesturnus rufusater) and South Island (SI) saddlebacks (P. carunculatus) differ in plumage and apparent vulnerability to introduced predators and, until recently, were considered subspecific. Here, we formally compare vocalisations, morphology and mtDNA between NI and SI saddlebacks. We show significant differences in male song and chatter calls which are fundamental saddleback vocalisations. A discriminant analysis correctly allocated 90% of birds based on significantly larger tarsus, weight and wing measurements in SI saddlebacks relative to NI saddlebacks. Finally, molecular data show c. 5% sequence divergence in mtDNA between the two groups. We concur with previous work elevating NI and SI saddleback to full species.
Introduction
There are both practical and theoretical reasons for defining and classifying species. Subsequently, the debate as to what constitutes a species is long running, often rancorous and, tellingly, shows no sign of conciliation (Townsend Peterson Citation1998; Coyne & Orr Citation2004; Price Citation2008). However, scientific progress and conservation management rely on objective and accurate definitions, despite the inherent challenges (Funk et al. Citation2002; Moritz Citation2002). The biological species concept (BSC) (Mayr Citation1969; Coyne & Orr Citation2004; Price Citation2008) has long been dominant in ornithology, but many viable alternatives have been proposed including the phylogenetic species concept (PSC; Cracraft Citation1983; McKitrick & Zink Citation1988), the evolutionary species concept (ESC; Wiley Citation1978; Weins Citation2004) and the general lineage concept (GLC; de Queiroz Citation1998, Citation1999).
Helbig et al. (Citation2002) suggested that differences among species concepts are primarily a matter of emphasis, but that the GLC encompassed many biologists' views of a species (i.e. species are population lineages that maintain their integrity in space and time relative to other lineages, are diagnosably different, reproductively isolated, share a common mate recognition and fertilisation system, and have their own independent evolutionary histories). The various species concepts lead to differing predictions about the outcome of future contact between species and Helbig et al. (Citation2002) proceeded to delimit and define species based on two criteria: are the taxa diagnosable (e.g. on the basis of plumage, vocalisations, morphology or DNA sequences)?; and are they likely to retain their genetic and phenotypic integrity in the future?
Recent ornithological taxonomy within New Zealand has largely adhered to the BSC (Oliver Citation1955; Heather & Robertson Citation1996; Higgins et al. Citation2006), with particular use of the subspecies concept to define geographically separated and distinct groups on the main islands (e.g. the North, South and Stewart Islands). However, the subspecies category itself is controversial (Rising Citation2007; Winker et al. Citation2007). This is understandable, particularly in continental systems where overlapping distributions of clinal species complicate rigorous definition of species limits (Patten & Unitt Citation2002; Cicero & Johnson Citation2006). However, the evolutionary processes influencing island species are often quite different, the potential for rapid change much more apparent (Rising Citation2007), and the distinction between species and subspecies, at least in comparison to continental systems, perhaps more definitive. Holdaway et al. (Citation2001) recognised this distinction in a recent revision of the breeding birds of New Zealand and argued for a reclassification of many New Zealand subspecies as good species, many of which were subsequently recognised as full species in the most recent checklist of the birds of New Zealand (Gill et al. Citation2010).
The New Zealand saddleback (Philesturnus) is a passerine in the family Callaeatidae (Holdaway et al. Citation2001; Shepherd & Lambert Citation2007). The saddleback was first described by Māori who called it either tīeke, an onomatopoeic name based on the birds' loud call, or tiaki meaning to guard or keep watch (Orbell Citation2003). The first European record of the South Island (SI) saddleback (P. carunculatus) was made in Queen Charlotte Sound in January 1770 while the North Island (NI) saddleback (P. rufusater) was first recorded in the Bay of Islands in 1772 (Oliver Citation1955; Watola Citation2007). The two forms have generally been considered subspecifically distinct since Mathews (Citation1943) synomised P. rufusater with P. carunculatus (Oliver Citation1955; Williams Citation1976; Turbott Citation1990; Heather & Robertson Citation1996; Higgins et al. Citation2006), but Holdaway et al. (Citation2001) cited earlier work, distinct plumage differences and disparity in body size in supporting full species status for the NI and SI birds. This has subsequently been recognised in the most recent checklist of the birds of New Zealand (Gill et al. Citation2010). Specifically, immature and juvenile SI saddlebacks have a brown plumage in contrast to the mostly black plumage of adult SI birds whereas immature and juvenile NI saddlebacks closely resemble adult NI birds (Higgins et al. Citation2006). Adult NI birds have a thin yellowish band at the front of the mantle that is absent in SI birds and the SI birds are larger than NI birds. There also appears to be differences in vulnerability to introduced predators, with NI saddleback both persisting and establishing in the presence of the non-native kiore or Polynesian rat (Rattus exulans) while the SI saddleback has not (Lovegrove Citation1996; Hooson & Jamieson Citation2003). Distinct differences in vocalisations are also apparent.
Advances in analytical methods, particularly of vocalisations and genetics, allow for careful appraisal of species limits in many birds (Townsend Peterson Citation1998; Rising Citation2007). Here, we formally compare NI and SI saddleback vocalisations, morphology and mtDNA, and assess the differences between these two taxa using the criteria described in Helbig et al. (Citation2002) for defining species limits.
Methods
Most of our NI saddleback data, particularly the vocalisation data, are drawn from all existing NI populations. However, the mtDNA data are from Hen Island only. The SI vocalisation and morphological data are from a single population on Ulva Island, and the mtDNA from Big Island, the source population for the saddleback that were reintroduced to Ulva. The two SI sites represent just two of 19 saddleback populations (Hooson & Jamieson Citation2003; Masuda et al. Citation2010). If the SI data we present are significantly different to all NI populations, then we think this comparison is sufficient for assessing distinctiveness between the two groups. Furthermore, for the purposes of these analyses we are only interested in differences between the NI and SI populations; variation within the NI saddleback data is discussed elsewhere (Parker et al. Citation2012).
Vocalisation recording and definition
NI saddleback vocalisations (n = 2513 male rhythmical songs from 1389 individuals; mean number of individuals recorded per island = 108.43 ± 19.71 [SEM]; mean number of songs per individual = 1.8 ± 0.7 and 257 chatter calls from 55 individuals; mean number of individuals recorded per population = 3.93 ± 0.30; mean number of calls per individual = 4.67 ± 0.52) were sampled by one to four researchers on each of the 13 established island populations (Hooson & Jamieson Citation2003; Parker Citation2008) on one to four trips of 5–15 days between December 2004 and November 2008. Observations were made across the entire area of most islands. However, only two areas were subsampled on Little Barrier Island reflecting two distinct translocation release locations and yielding a total of 14 sample populations. Vocalisations were recorded using Sony Hi MD MZ-NH700 mini discs, Sennheiser K6 recording modules, M66 microphones and Rycote Softy wind guards mounted on Rycote pistol grips. Vocalisations were defined as loud chatter calls (given by both sexes), sexually dimorphic quiet calls and male rhythmical songs (Jenkins Citation1977). SI saddleback vocalisations (n = 99 male songs from 25 individuals with a mean of 3.96 ± 0.65 [SEM] songs per individual and 80 chatter calls from 27 individuals with a mean of 2.96 ± 0.47 calls per individual) were recorded on Ulva Island from October to December 2004 using a Sony TCD-D8 digital audio tape recorder, Sennheiser MKH 815T microphone and Rycote windshield following the method of Ludwig and Jamieson (Citation2007). NI saddleback calls were downloaded and converted to WAV. format using Sony SonicStage software (Sony Corporation 2001–2007) with 16 bit sampling precision and a 44 kHz sampling rate. SI saddleback vocalisations were directly recorded in the same format (Ludwig & Jamieson Citation2007). SI saddleback vocalisations were defined as chatter calls, non-sexually dimorphic quiet calls and male specific phrases (Ludwig & Jamieson Citation2007). Subsequent analysis was restricted to a comparison of NI saddleback male rhythmical song and the analogous SI saddleback male specific song and a separate comparison of NI and SI saddleback chatter calls.
NI and SI saddleback vocalisations are easily distinguished both aurally and visually on a spectrogram by researchers (Ludwig & Jamieson Citation2007; Parker et al. Citation2010) and all recordings were inspected (FFT = 256, Hann window, 5.8 ms, 50% overlap) and categorised as chatter calls, NI saddleback MRS or SI saddleback male specific phrases using Raven 1.2.1 (Cornell Lab of Ornithology, Ithaca, NY, USA). NI saddleback male rhythmical song consists of highly stereotyped repeated phrases sung by male birds in territorial displays (Jenkins Citation1977) (A). SI saddlebacks sing male specific phrases comparable to NI saddleback male rhythmical song (B) but do not show the high degree of stereotypy observed in NI saddleback and instead deliver songs using partial and full phrases in a mixed mode and random order (Ludwig & Jamieson Citation2007). Given that both NI and SI birds use phrases to form their songs, we used a single phrase from each male song as the unit of song comparison (A–B).
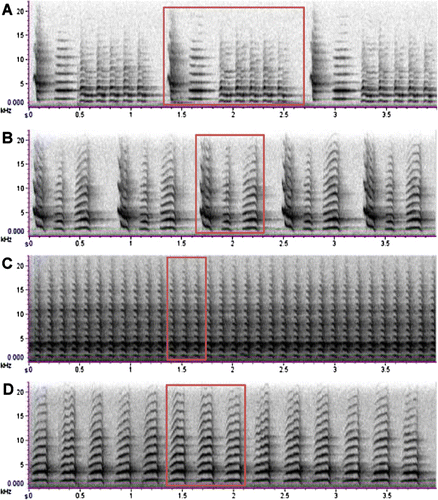
NI and SI chatter calls are similar (C–D) both consisting of repeated phrases delivered in widely varying lengths depending on the social context of their delivery (Jenkins Citation1977; Ludwig & Jamieson Citation2007). Therefore, for our unit of comparison we measured three consecutive chatter phrases within a single chatter call (C–D).
Spectral, temporal and amplitude measurements of individual NI and SI phrases were taken in Sound Analysis Pro v1.02 (Tchernichovski et al. Citation2004) using the manual segmentation procedure with a 200 Hz high pass filter (data window = 9.27 ms, advance window = 1.36 ms, spectrum range = 11 180 Hz, Fast Fourier Transform window = 1024 samples, tapers = 2, bandwidth parameter = 1.5). Where multiple songs or chatter calls were recorded from a single individual, the variables were averaged, giving a single value for each variable for each bird recorded. The following spectral, temporal and amplitude variables were used for analysis (see Tchernichovski et al. [Citation2004] and Parker et al. [Citation2012] for variable definitions): duration; mean amplitude modulation; and both the mean and the variance of pitch, entropy, frequency modulation, goodness of pitch and mean frequency. Two datasets were created from this procedure, one consisting of NI saddleback male rhythmical song and SI saddleback male specific phrases and one consisting of NI and SI chatter calls.
Spectral, temporal and amplitude variables were examined for normality and the following variables were transformed to ln(x) + 0.01 to remove skewness for all male songs and chatter calls: duration; mean pitch; variance in pitch; and variance in goodness of pitch. The following variables were transformed for analysis of male songs, but not for chatter calls: mean goodness of pitch; mean frequency; variance in frequency; and variance in entropy.
Heterogeneity in spectral, temporal and amplitude characteristics of male songs and chatter calls among NI saddleback populations and Ulva Island SI saddleback was examined using the distance-based test of multivariate dispersions (variance) (PERMDISP, 9999 permutations; Anderson Citation2006) following standardisation to z-scores (i.e. normalisation). Spectral, temporal and amplitude variables were further analysed using island population means and the relationships among these populations were examined using principal components analysis (PCA).
Morphological measurements
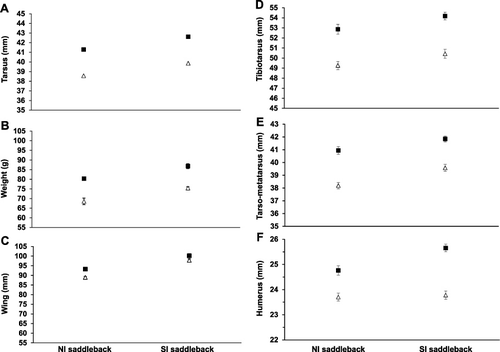
NI saddlebacks (n = 32 males and 23 females) were captured in mist nets (Hen, Cuvier, Tiritiri Matangi and Mokoia Islands) or in roost boxes (Tiritiri Matangi Island) between August 2005 and August 2006. Tarsus length (intra-articular notch at the proximal end of the tarsometatarsus to the middle of the midtarsal articulation; see Taylor & Jamieson Citation2007) was measured twice to the nearest ± 0.01 mm with vernier callipers. Wing length (maximum unflattened wing chord) was measured to the nearest ± 0.5 mm with a wing rule and weight was measured to the nearest ± 0.5 g with a Pesola spring scale. Sex and age were determined by tarsus length and plumage respectively after Jenkins & Veitch (Citation1991). All measurements were taken by KAP.
SI saddlebacks (n = 20 males and 20 females) were captured in mist nets on Ulva Island in April 2009 for subsequent translocation to Orokonui Ecosanctuary (Masuda et al. Citation2010). Identical measurements were taken but tarsus length was measured three times to the nearest ± 0.1 mm, wing length to the nearest ± 1 mm and weight to the nearest ± 1 g. Sex was determined using methodology outlined in Taylor and Jamieson (Citation2007) and age after Heather and Robertson (Citation1996). All measurements were taken by B.M. Masuda (Otago University, Dunedin, New Zealand).
The NI saddleback tarsus, wing and weight measurements were rounded to the same scale as the SI saddleback data and then all data were examined for normality. We first calculated a resemblance matrix based on Euclidean distance and then checked the NI and SI data for homogeneity of multivariate dispersion (variance) (PERMDISP, 9999 permutations; Anderson Citation2006) followed with a non-parametric multivariate analysis of variance (PERMANOVA, 9999 permutations; Anderson Citation2001). We then did a classical discriminant analysis using canonical analysis of principal coordinates (CAP, 9999 permutations; Anderson & Robinson Citation2003; Anderson & Willis Citation2003). These analyses were repeated to examine the data by origin (NI or SI) and sex. Finally, we examined each variable individually using univariate two-way ANOVA with origin and sex as the grouping variables with Tukey's HSD and Bonferroni-adjusted significance levels.
In addition to measurements from live birds we compared the greatest length ± 0.01 mm from complete skeletons held in the Museum of New Zealand Te Papa Tongarewa collection (see Table S1). The measured bones were the tibiotarsus, tarsometatarsus, femur, humerus and ulna (measured by P. Scofield; n = seven NI females, five NI males, six SI females and eight SI males). Sex was determined either from museum labels (46%, n = 26) or was inferred from measurements (54%, n = 26).
All vocalisation and morphological multivariate analyses were done using the PRIMER v6 computer program (Clarke & Gorley Citation2006) with the PERMANOVA+ add-on package (Anderson et al. Citation2008). Univariate analyses were done using Statistica v6 (STATISTICA Citation2002).
Mitochondrial DNA
Blood from four NI Saddlebacks (Hen Island) and five SI Saddlebacks (Big Island) was collected by the authors. Two NI kōkako (Callaeas cinerea wilsoni) samples were provided by I. Hogg (University of Waikato) and were collected as part of another study (Hudson et al. Citation2000). All blood samples were stored in Seutin buffer (Seutin et al. Citation1991). Five microlitres of blood in buffer was added to 400 µl of 5% Chelex solution (BioRad) containing 40 µg Proteinase K (Roche) and incubated overnight at 65 °C, followed by 10 min at 95 °C.
Approximately 850 bp of control region was amplified using primers L437 (5′ CTCACGAGAACCGAGCTACT 3′) and H1248 (5′ CATCTTCAGTGTCATGCT 3′) (Tarr Citation1995). Each 20 µl PCR contained 0.5 µM of each primer, 0.8 mM dNTPs, 1.5 mM MgCl2 and 0.5 U of BioTaq (Bioline). PCR amplifications were performed using an Eppendorf Mastercycler ep gradient S and consisted of 94 °C for 120 s, followed by 35 cycles of 94 °C for 30 s, 50 °C for 30 s and 72 °C for 60 s, with a final extension step of 72 °C for 240 s. Amplified DNA was purified using a High Pure PCR Purification Kit (Roche) and quantified using a Nanodrop ND-1000 spectrophotometer. Templates were sequenced on an ABI 3730 DNA Analyzer using primer L437 and an internal primer, L-DomIII (5′ CTCACACTTTGCCCTGATGC 3′; Double & Murphy Citation2000).
Sequences were edited and aligned along with additional sequences from GenBank using MUSCLE in Geneious v 6.0.3 (Biomatters, available from http://www.geneious.com) and adjusted manually as necessary, then compared to the NCBI database using BLAST to search for genetic matches (Altschul et al. Citation1990). Phylogenetic analyses were performed using PAUP* v 4b10 (Swofford Citation2002). Neighbour joining (NJ) trees were constructed using general time-reversible (GTR) corrected distances and 1000 bootstrap replicates were conducted to assess support for each of the nodes. Distance calculations were performed in Geneious v6.0.3. Sequences have been submitted to Genbank (Accession numbers HQ606050–HQ 606060).
Results
Song
Highly significant differences in multivariate dispersion (variance) of spectral, temporal and amplitude characteristics (PERMDISP, F = 8.85, P = 0.0001, 9999 permutations) are evident when male specific calls from SI saddleback on Ulva Island are compared with NI saddleback male rhythmical song (A). Pairwise comparisons reveal that SI saddleback on Ulva Island show significantly less spectral, temporal and amplitude diversity in song than any NI saddleback population (P ≤ 0.0024).
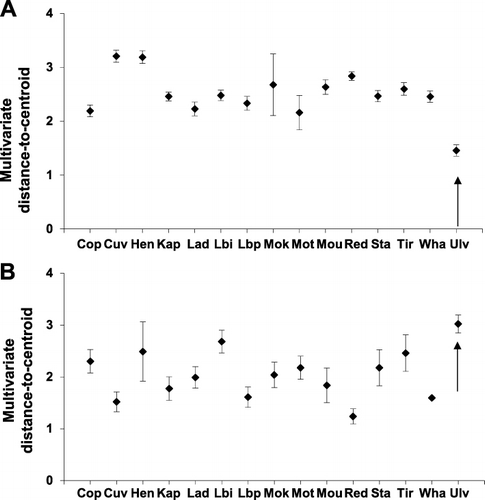
In contrast, chatter calls from SI saddleback on Ulva Island show significantly greater multivariate dispersion of spectral, temporal and amplitude characteristics than most NI saddleback populations (PERMDISP, F = 5.88, P = 0.0001, 9999 permutations; B). Pairwise comparisons show significant differences (P ≤ 0.045) among five of 14 NI saddleback populations (Cuvier, Lady Alice, Little Barrier Pohutukawa Flat, Mokoia, Red Mercury), but no difference among the remaining nine populations.
When examined on a PCA ordination the differences between all NI saddleback populations and SI saddleback from Ulva Island in both song and chatter calls are apparent (). SI saddleback songs from Ulva Island show consistently higher PC1 and PC2 values relative to NI saddleback populations (A; ) and consistently lower PC1 values and generally higher PC2 values for chatter calls relative to NI saddleback populations (B; ).
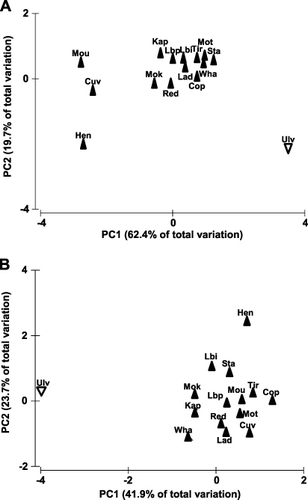
Table 1 Principal component factor loadings for the analysis of spectral, temporal and amplitude variables from male specific song and chatter calls from island populations of NI and SI saddleback.
Morphology
There was no significant difference in multivariate dispersion (variance) of the morphological variables from live birds (PERMDISP, F = 0.51, P = 0.48; NI saddleback mean dispersion = 8.18 ± 0.60 [SEM, n = 55; SI saddleback = 7.48 ± 0.59, n = 40) or bone measurements (PERMDISP, F = 0.06, P = 0.79; NI saddleback mean dispersion = 2.60 ± 0.38, n = 12; SI saddleback = 2.72 ± 0.28, n = 14), but there was a highly significant difference in multivariate location between NI and SI saddleback for measurements from live birds (PERMANOVA, pseudo-F1,93 = 26.25, P = 0.0001) and those from bones (PERMANOVA, pseudo-F1,24 = 4.61, P = 0.03). A single canonical axis (δ12 = 0.56) was calculated in the discriminant analyses (CAP) as there are only two groups. The leave-one-out allocation success was high for measures from live birds with 90.52% (86/95) of samples assigned to the correct group, but lower for bone measurements at 76.92% (20/26). NI saddlebacks (live birds 92.73%, 51/55; bone measurements 83.33%, 10/12) were correctly assigned at a slightly higher proportion than SI saddlebacks (live birds 87.50%, 35/40; bone measurements 71.43%, 10/14). In permutation tests the trace statistics (0.56) and the first squared canonical correlations (0.56) were highly significant (P = 0.0001) (the two statistics are identical because there are only two groups in each CAP; Anderson et al. Citation2008).
There was a significant difference in multivariate dispersion (variance) when the data from live birds were examined by origin and sex (PERMDISP, F = 3.79, P = 0.01) with female NI (6.88 ± 1.02, n = 23) and male SI saddleback (6.21 ± 0.71, n = 20) showing greater multivariate dispersion than male NI (4.32 ± 0.52, n = 32) and female SI saddleback (4.15 ± 0.42, n = 20) but not for bone measurements (PERMDISP, F = 3.57, P = 0.10; female NI (1.63 ± 0.16, n = 7), male NI saddleback (1.18 ± 0.16, n = 5), female SI (0.90 ± 0.13, n = 6), male SI saddleback (1.31 ± 0.17, n = 8). PERMANOVA is sensitive to differences in dispersion (variance) among groups but Anderson et al. (Citation2008) suggest that it is robust to some heterogeneity. Furthermore, examination of the data in an unconstrained non-metric multidimensional scaling (MDS) ordination suggests that there are differences in location as well as dispersion (A). Therefore, we conducted a further test of multivariate location by sex and origin for both live birds and bones which revealed a significant difference between groups (live birds, PERMANOVA, pseudo-F3,91 = 45.21, P = 0.0001; bones, PERMANOVA, pseudo-F3,22 = 32.30, P = 0.0001). The first (live birds, δ12 = 0.77; bones δ12 = 0.87) and second canonical (live birds, δ12 = 0.51; bones δ12 = 0.54) axes in the discriminant analyses (CAP) clearly separate the data by sex and origin and the leave-one-out allocation success was reasonably high for live birds (84.21%, 80/95; B) but lower for bone measures (69.23%, 18/26; D). NI males (90.63%, 29/32) and NI females (86.96%, 20/23) were correctly assigned more often than male SI (75%, 15/20) and female SI birds (80%, 16/20) from live measurements and NI (85.71%, 6/7) and SI females (83.33, 5/6) were correctly assigned more often than NI (60%, 3/5) and SI males (50%, 4/8) from bones. In permutation tests, the trace statistics (live birds, 1.28; bones, 1.71) and the first squared canonical correlations (live birds, 0.77; bones, 0.89) were highly significant (P = 0.0001).
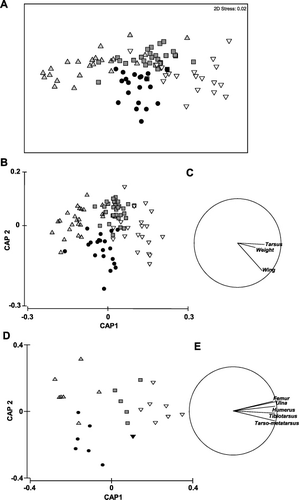
Univariate analyses showed there are significant differences (uncorrected P values are shown) in tarsus (F1,91 = 51.92, P < 0.00001) between live NI and SI birds with Tukey's HSD showing significant differences between male NI, female NI, male SI and female SI birds (P < 0.0002; A). There are also significant differences in weight (F1,91 = 32.18, P < 0.00001) with Tukey's HSD showing significant differences between all four groups (P ≤ 0.0008; B) and in wing measurements (F1,91 = 149.77, P < 0.00001) with Tukey's HSD showing significant differences between all groups (P < 0.0002) except male and female SI birds (P = 0.07; C). For bone measurements, there were significant differences between NI and SI birds for tibiotarsus (F1,22 = 8.02, P = 0.0097) with Tukey's HSD showing significant differences between (P ≤ 0.006; D) but not among sexes (P > 0.18; D). There were also significant differences in tarsometatarsus (F1,22 = 17.48, P = 0.0004) with Tukey's HSD showing significant differences between all groups (P ≤ 0.007; E) except NI and SI males (P > 0.13; E) and in humerus (F1,22 = 8.34, P = 0.0086) with Tukey's HSD showing significant differences between all groups (P ≤ 0.001; F) except NI and SI females (P > 0.98; F). There were no significant differences between NI and SI birds for femur length (F1,22 = 0.4, P = 0.53; NI male = 29.34 ± 0.21 [SEM], NI female = 27.9 ± 0.18; SI male = 29.34 ± 0.21, SI female = 27.9 ± 0.18) or ulna length (F1,22 = 3.67, P = 0.068; NI male = 26.26 ± 0.14, NI female = 25.19 ± 0.12; SI male = 26.79 ± 0.11, SI female = 25.15 ± 0.13). The Bonferroni adjusted experiment-wise significance level for conducting eight simultaneous tests was 0.05/8 = 0.006.
mtDNA
We obtained 770 bp of sequence for NI saddleback, 768 bp for SI saddleback and 804 bp for NI kōkako. BLAST searches confirmed the sequences to be control region with closest matches to NI and SI kōkako (C. c. cinerea). There was no within species variation for any of the taxa. Sequences from the present study were aligned with South Island kōkako (AF433181 and AF433200) and huia (Heteralocha acutirostris, AF433205). The GenBank sequences were approximately 400 bp shorter than those in the present study. Our additional sequence was part of domain II of the control region, which is significantly less variable than the other two domains (Baker & Marshall Citation1997). To avoid bias we analysed the two sequence regions separately and indels were treated as single events where all other sequences were identical at those positions.
North and South Island saddleback are sister taxa clearly differentiated from each other and from kōkako on a NJ tree (see Fig. S1). shows a distance matrix, with the number of base differences as well as percentage sequence divergence. We saw 2.38% divergence in the first sequence region and 5.09% in the second sequence region between the two saddleback subspecies, which equates to 10 and 20 base differences, respectively. This is comparable to the 4.71% (second sequence region only) mean divergence (17–21 bases) between NI and SI kōkako. The mean level of divergence between saddlebacks and kōkako was 9.44%/14.53% (38–41 bases and 53–60 bases, respectively). These data contrast to relatively low levels of divergence (0.6%) among isolated populations of NI kōkako (Murphy et al. Citation2006) and the complete lack of variation we saw in NI and SI saddlebacks.
Table 2 Within and between species or subspecies sequence divergence. The absolute number of nucleotide differences are shown first and percentage sequence divergence second. Data for the first 420 aligned bases are shown above the diagonal and the second 393 aligned bases below.
Discussion
The vocalisation, morphological and mtDNA data presented here, along with plumage differences (Higgins et al. Citation2006) and apparent differences in vulnerabilities to introduced predators (Lovegrove Citation1996; Hooson & Jamieson Citation2003), consistently show large and statistically significant differences between NI and at least one SI saddleback population.
Helbig et al.'s (Citation2002) criteria demand differentiation in at least one of three criteria: 1. individuals of at least one age/sex class can be distinguished from individuals of the same age/sex class of the other taxa by at least one qualitative difference; 2. at least one age/sex class is separated by a complete discontinuity in at least one continuously varying character from the same age/sex class; and 3. individuals of at least one age/sex class can be clearly distinguished from individuals of the same age/sex class of all other taxa by a combination of two or three functionally independent characters. The species level separation of NI and SI saddleback clearly fulfil Helbig et al.'s (Citation2002) first criteria due to completely distinct plumage in both juveniles (which resemble adults in the NI, but in the SI have a distinct brown plumage) and adults (yellowish band present in the NI and absent in the SI; Higgins et al. Citation2006).
Close examination of the vocalisation characters did not show complete discontinuity, thereby Helbig et al.'s (Citation2002) second criterion is not fulfilled. However, there is virtually no overlap in multivariate dispersion (A,–B) for two of the three main categories of saddleback vocalisations (the third category, quiet calls, are not included in the analyses presented here), and there are clear differences in use of multivariate space (A–B, ) when NI island populations are compared to the SI population on Ulva Island. The observed differences are also immediately apparent when listening to NI and SI saddleback vocalisations. In addition, we question whether complete discontinuity in song characters is necessary for assessing differences between taxonomic groups, particularly given the very similar vocalisations of pairs of species within many genera (e.g. Turdus spp.). Further analyses quantifying song characteristics relative to the closely related NI kōkako, the only other extant wattlebird, might elucidate the significance of differences in song between NI and SI saddleback.
Complete discontinuity was not evident in the morphological characters compared here, although relatively good discrimination in discriminant analysis (B, D) and significant mean differences (A–F) were observed. However, we treat these differences with caution as a trend for increasing body size with increasing latitude is apparent in many species (Bergmann Citation1847; Blackburn et al. Citation1999), including most New Zealand bird species (Higgins et al. Citation2006). We also note the relatively small sample sizes for morphological data compared here, particularly for the bone measurements from museum collections.
The percentage divergence in mtDNA presented here is similar to NI/SI levels of divergence in other bird species such as kōkako (Murphy et al. Citation2006) and blue duck (Hymenolaimus malacorhynchos) (Robertson et al. Citation2007), and is thought to date back about 0.8–0.9 million years (Murphy et al. Citation2006). Divergence times between NI and SI avifauna need to be treated cautiously, but are likely to be within the correct order of magnitude (Baker et al. Citation1995; Burbidge et al. Citation2003), and therefore provide ample time for NI and SI saddleback to diverge in vocalisations, morphology and behaviour. The level of sequence divergence (5.6%) between NI and SI saddlebacks is sufficient to support elevation to full species status and is similar to that used by Miller and Lambert (Citation2006) to assign species status to the previously subspecific North Island (Petroica longipes) and South Island robins (P. australis) in New Zealand. It is also similar to levels of divergence seen in a meta-analysis of 643 North American bird species (Kerr et al. Citation2007). Hence, we interpret these mtDNA data as evidence of independent evolutionary histories and support for Helbig et al.'s (Citation2002) second criteria for diagnosability.
Helbig et al. (Citation2002) require an assessment of the likelihood that taxa will retain their genetic and phenotypic integrity into the future to assign a rank of species. In contrast to allopatric taxa, such as the NI and SI saddleback, the reproductive isolation of sympatric and parapatric taxa can be assessed through direct observation. Therefore, the likelihood that the historically allopatric NI and SI forms will remain distinct becomes hypothetical. The role of plumage differences between NI and SI saddleback is unknown, although we note that in performing the bow fan warble display (Higgins et al. Citation2006) male NI birds present their mantle to either their mate or a territorial foe. We also note that SI saddleback can breed whilst in their juvenile plumage. Bird vocalisations play a critical role in species recognition, mate choice and resource defence (Catchpole & Slater Citation1995), and likely influence speciation (Grant & Grant Citation1997; Slabbekoorn & Smith Citation2002). Therefore, playback experiments might also help elucidate the degree of reproductive isolation, particularly as NI saddleback can discriminate between songs that are much more similar than the differences observed between NI and SI songs (Parker et al. Citation2010). Finally, Helbig et al. (Citation2002) state that allopatric taxa can be assigned species rank if several discrete or continuously varying characters are diagnosable and the sum of character differences corresponds to or exceeds that seen in closely related sympatric species. This comparison is not possible for saddleback as there are no extant closely related sympatric species aside from the NI kōkako.
Saddlebacks are reproductively isolated because of habitat specificity to forest and scrubland habitats and an apparent inability to disperse across gaps greater than 250 m (Weins Citation2004; Higgins et al. Citation2006). This, along with human mediated landscape changes, means that NI and SI saddleback are extremely unlikely to ever meet in sympatry. Experimental mixing of NI and SI birds would be uninformative, as even distantly related birds can successfully hybridise (Grant & Grant Citation1997; Allendorf et al. Citation2001), and inadvisable given that the two forms of saddleback comprise evolutionary significant units for conservation purposes (Moritz Citation1994, Citation2002). Therefore, given the vocalisation, morphological and mtDNA differences we describe here we concur with Holdaway et al. (Citation2001) and recommend elevating the NI and SI saddleback to full species status. The species commonly called the South Island saddleback shall be known as Philesturnus carunculatus and the North Island saddleback shall be known as Philesturnus rufusater.
Supplementary files
Supplementary file 1: Table 1. Complete North Island and South Island saddleback skeletons measured from the Museum of New Zealand Te Papa Tongarewa collection.
Supplementary file 2: Figure 1. A neighbour joining phylogram estimated from 813 aligned nucleotides of the control region. The phylogram has a basal polytomy, which is consistent with previous studies (Shepherd & Lambert Citation2007) that were unable to resolve the relationships between these taxa. Bootstrap values > 50% are shown, based on 1000 replicates.
Figure 1. A neighbour joining phylogram estimated from 813 aligned nucleotides of the control region.
Download PDF (75.2 KB)Table 1. Complete North Island and South Island saddleback skeletons measured from the Museum of New Zealand Te Papa Tongarewa collection.
Download MS Word (14.1 KB)Acknowledgements
This research was part of KAP's PhD research and was funded by a Marsden Grant through the Royal Society of New Zealand to DHB. IGJ's research on conservation genetics of SI saddleback was funded by Department of Conservation (contract no. 3576), Landcare Research (contract no. C09X0503) and the University of Otago. We thank N. Adams, D. Alverez, J. Barr, P. Barrow, M. Booth, I. Castro, T. Cope, M. Delany, S. Heiss-Dunlop, M. Duplain, B. Evans, R. Gardner-Gee, S. Graham, R. Griffiths, M. Harrison, J. Hilton, W. Ji, H. Lindsay, L. Ortiz Catedral, G. Parker, S. Stoddard, R. Thorogood, R.J. Thorogood, B. Zeisemann and New Zealand Department of Conservation staff for assistance in collecting NI saddleback data. Ngāti Wai, Ngāti Manuhiri, Ngāti Rehua, Ngāti Paoa, Te Kawerau a Maki, Ngāti Hei, Ngāti Tamaterā, the Mokoia Trust Board, Ngāti Awa and Te Āti Awa all kindly provided access to motu (islands) within their rohe (areas). B. Masuda provided morphological data on SI saddlebacks, M. Kennedy and D.Winter assisted with the genetic analysis and I. Hogg and the New Zealand Department of Conservation Kōkako Recovery Group provided kōkako blood samples for the sequencing analysis. Fieldwork and bird capture was conducted under permit from the New Zealand Department of Conservation and the Massey and Otago University Animal Ethics Committees. This manuscript benefited from constructive comments provided by J. Banks, T. Worthy and an anonymous reviewer.
References
- Allendorf FW, Leary RF, Spruell P, Wenburg JK 2001. The problems with hybrids: setting conservation guidelines. Trends in Ecology & Evolution 16: 613–622. 10.1016/S0169-5347(01)02290-X
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ 1990. Basic local alignment search tool. Journal of Molecular Biology 215: 403–410.
- Anderson MJ 2001. A new method for non-parametric multivariate analysis of variance. Austral Ecology 26: 32–46.
- Anderson MJ 2006. Distance based tests for homogeneity of multivariate dispersions. Biometrics 62: 245–253. 10.1111/j.1541-0420.2005.00440.x
- Anderson MJ, Robinson J 2003. Generalized discriminant analysis based on distances. Australian and New Zealand Journal of Statistics 45: 301–318. 10.1111/1467-842X.00285
- Anderson MJ, Gorley RN, Clarke KR 2008. PERMANOVA+ for PRIMER: guide to software and statistical methods. Plymouth, PRIMER-E Ltd. 214 p.
- Anderson MJ, Willis TJ 2003. Canonical analysis of principle coordinates: a useful method of constrained ordination for ecology. Ecology 84: 511–525. 10.1890/0012-9658(2003)084[0511:CAOPCA]2.0.CO;2
- Baker AJ, Daugherty CH, Colbourne R, McLennan JL 1995. Flightless brown kiwis of New Zealand possess extremely subdivided population structure and cryptic species like small mammals. Proceedings of the National Academy of Sciences of the United States of America 92: 8254–8258. 10.1073/pnas.92.18.8254
- Baker AJ, Marshall HD 1997. Mitochondrial control region sequences as tools for understanding evolution. In: Mindell DP ed Avian molecular evolution and systematics. San Diego, Academic Press. Pp. 51–82.
- Bergmann C 1847. Über die Verhältnisse der Wärmeokonomie der Thiere zu iher Grösse. [On the relations of the heat economy of the animals to their size] Göttinger Studien 3: 595–708.
- Blackburn TM, Gaston KJ, Loder N 1999. Geographic gradients in body size: a clarification of Bergmann's Rule. Diversity and Distributions 5: 165–174. 10.1046/j.1472-4642.1999.00046.x
- Burbidge ML, Colbourne RM, Robertson HA, Baker AJ 2003. Molecular and other biological evidence supports the recognition of at least three species of brown kiwi. Conservation Genetics 4: 167–177. 10.1023/A:1023386506067
- Catchpole CK, Slater PJB 1995. Bird song: biological themes and variations. Cambridge, Cambridge University Press. 248 p.
- Cicero C, Johnson NK 2006. Diagnosability of subspecies: lessons from sage sparrows (Amphispiza belli) for analysis of geographic variation in birds. Auk 123: 266–274. 10.1642/0004-8038(2006)123[0266:DOSLFS]2.0.CO;2
- Clarke KR, Gorley RN 2006. PRIMER v6: user manual/tutorial. Plymouth, PRIMER-E Ltd.
- Coyne JA, Orr HA 2004. Speciation. Massachusetts, Sinauer Associates. 545 p.
- Cracraft J 1983. Species concepts and speciation analysis. Current Ornithology 1: 159–187.
- de Queiroz K 1998. The general lineage concept of species, species criteria, and the process of speciation. A conceptual unification and terminological recommendations. In: Howard DJ, Berlocher SH eds. Endless forms: species and speciation. Oxford, Oxford University Press. Pp. 57–75.
- de Queiroz K 1999. The general lineage concept of species and the defining properties of the species category. In: Wilson RA ed Species, new interdisciplinary essays. Cambridge, MA, MIT Press. Pp. 49–89.
- Double M, Murphy S 2000. Genetic variation within and among populations of North Island kokako. Department of Conservation Science & Research Internal Report 176. Wellington, New Zealand, Department of Conservation.
- Funk VA, Sakai AK, Richardson K 2002. Biodiversity: the interface between systematics and conservation. Systematic Biology 51: 235–237. 10.1080/10635150252899743
- Gill BJ, Bell BD, Chambers GK, Medway DG, Palma RL, Scofield RP, et al. 2010. Checklist of the birds of New Zealand, Norfolk, and Macquarie Islands, and the Ross dependency, Antarctica. 4th edition. Wellington, Ornithological Society of New Zealand and Te Papa Press. 500 p.
- Grant PR, Grant BR 1997. Genetics and the origin of bird species. Proceedings of the National Academy of Sciences of the United States of America 94: 7768–7775. 10.1073/pnas.94.15.7768
- Heather B, Robertson H 1996. The field guide to the birds of New Zealand. Auckland, Viking, Penguin Books. 432 p.
- Helbig AJ, Knox AG, Parkin DT, Sangster G, Collinson M 2002. Guidelines for assigning species rank. Ibis 144: 518–525. 10.1046/j.1474-919X.2002.00091.x
- Higgins PJ, Peter JM, Cowling SJ, eds. 2006. Handbook of Australian, New Zealand and Antarctic birds: boatbill to starlings. Melbourne Oxford University Press. 1984 p.
- Holdaway RN, Worthy TH, Tennyson AJD 2001. A working list of breeding bird species of the New Zealand region at first human contact. New Zealand Journal of Zoology 28: 119–187. 10.1080/03014223.2001.9518262
- Hooson S, Jamieson IG 2003. The distribution and current status of New Zealand Saddleback Philesturnus carunculatus. Bird Conservation International 13: 79–95. 10.1017/S0959270903003083
- Hudson QJ, Wilkins RJ, Waas JR, Hogg ID 2000. Low genetic variability in small populations of New Zealand kokako (Callaeas cinerea wilsoni). Biological Conservation 96: 105–112. 10.1016/S0006-3207(00)00057-4
- Jenkins PF 1977. Cultural transmission of song patterns and dialect development in a free-living bird population. Animal Behaviour 25: 50–78.
- Jenkins PF, Veitch CR 1991. Sexual dimorphism and age-determination in the North Island Saddleback (Philesturnus carunculatus rufusater). New Zealand Journal of Zoology 18: 445–450. 10.1080/03014223.1991.10422851
- Kerr KCR, Stoeckle MY, Dove, CJ, Weigt LA, Francis CM, Hebert PDN 2007. Comprehensive DNA barcode coverage of North American birds. Molecular Ecology Notes 7: 535–543. 10.1111/j.1471-8286.2007.01670.x
- Lovegrove TG 1996. Island releases of saddlebacks Philesturnus carunculatus in New Zealand. Biological Conservation 77: 151–157. 10.1016/0006-3207(96)00006-7
- Ludwig K, Jamieson IG 2007. Phrase types, repertoire size and repertoire overlap in the South island saddleback (Philesturnus carunculatus carunculatus). Notornis 54: 201–213.
- Masuda BM, Smith ED, Jamieson IG 2010. Assessment of protocols and best-practise techniques learned during a translocation of South Island saddlebacks Philesturnus carunculatus from Ulva Island to Orokonui Ecosanctuary, New Zealand Conservation Evidence 7: 69–74.
- Mathews G 1943. Notes on New Zealand Birds. Emu 43: 243–247. 10.1071/MU943243
- Mayr E 1969. Principles of systematic zoology. New York, McGraw-Hill. 428 p.
- McKitrick MC, Zink RM 1988. Species concepts in ornithology. Condor 90: 1–14. 10.2307/1368426
- Miller HC, Lambert DM 2006. A molecular phylogeny of New Zealand's Petroica (Aves: Petroicidae) species based on mitochondrial DNA sequences. Molecular Phylogenetics and Evolution 40: 844–855. 10.1016/j.ympev.2006.04.012
- Moritz C 1994. Defining “Evolutionary Significant Units” for conservation. Trends in Ecology & Evolution 9: 373–375. 10.1016/0169-5347(94)90057-4
- Moritz C 2002. Strategies to protect biological diversity and the evolutionary processes that sustain it. Systematic Biology 51: 238–254. 10.1080/10635150252899752
- Murphy SA, Flux IA, Double MC 2006. Recent evolutionary history of New Zealand's North and South Island kokako (Callaeas cinerea) inferred from mitochondrial DNA sequences. Emu 106: 41–48. 10.1071/MU05007
- Oliver WRB 1955. New Zealand birds. 2nd Edition. Wellinton, A.H. and A.W. Reed. 661 p.
- Orbell M 2003. Birds of Aotearoa. A natural and cultural history. Auckland, Reed Publishing (NZ) Ltd. 198 p.
- Parker KA 2008. Translocations: providing outcomes for wildlife, resource managers, scientists, and the human community. Restoration Ecology 16: 204–209. 10.1111/j.1526-100X.2008.00388.x
- Parker KA, Anderson MJ, Jenkins PF, Brunton DH 2012. The effects of translocation-induced isolation and fragmentation on the cultural evolution of bird song. Ecology Letters 15: 778–785. 10.1111/j.1461-0248.2012.01797.x
- Parker KA, Hauber ME, Brunton DH 2010. Contemporary cultural evolution of a conspecific recognition signal following serial translocations. Evolution 64: 2431–2441.
- Patten MA, Unitt P 2002. Diagnosability versus mean differences of sage sparrow subspecies. Auk 119: 26–35.
- Price T 2008. Speciation in birds. Greenwood Village, CO, Roberts and Company. 470 p.
- Rising D 2007. Named subspecies and their significance in contemporary ornithology. In: Cicero C, Remsen JV eds. Festschrift for Ned K. Johnson: geographic variation and evolution in birds. Washington, DC, The American Ornithologists' Union. Pp. 45–54.
- Robertson BC, Steeves TE, McBride KP, Goldstien SJ, Williams M, Gemmell NJ 2007. Phylogeography of the New Zealand blue duck (Hymenolaimus malacorhynchos): implications for translocation and species recovery. Conservation Genetics 8: 1431–1440. 10.1007/s10592-007-9294-2
- Seutin G, White BN, Boag PT 1991. Preservation of avian blood and tissue samples for DNA analysis. Canadian Journal of Zoology-Revue Canadienne De Zoologie 69: 82–90. 10.1139/z91-013
- Shepherd LD, Lambert DM 2007. The relationships and origins of the New Zealand wattlebirds (Passeriformes, Callaeatidae) from DNA sequence analyses. Molecular Phylogenetics and Evolution 43: 480–492. 10.1016/j.ympev.2006.12.008
- Slabbekoorn H, Smith TB 2002. Bird song, ecology and speciation. Philosophical Transactions of the Royal Society B-Biological Sciences 357: 493–503. 10.1098/rstb.2001.1056
- STATISTICA 2002. v. 6.0 [software]. Tulsa-OK, USA, Statsoft Inc.
- Swofford DL 2002. PAUP* 4.0b10. Sunderland, MA, Sinauer Associates.
- Tarr CL 1995. Primers for amplification and determination of mitochondrial control-region sequences in oscine passerines. Molecular Ecology 4: 527–530. 10.1111/j.1365-294X.1995.tb00251.x
- Taylor SS, Jamieson IG 2007. Determining sex of South Island saddlebacks (Philesturnus carunculatus carunculatus) using discriminant function analysis. Notornis 54: 61–64.
- Tchernichovski O, Swigger D, Mitra P 2004. Sound Analysis Pro user manual. CCNY, New York. 134 p.
- Townsend Peterson A 1998. New species and new species limits in birds. Auk 115: 555–558. 10.2307/4089405
- Turbott EG ed. 1990. Checklist of the birds of New Zealand and the Ross dependency, Antarctica. Auckland, Random Century. 247 p.
- Watola GV 2007. New Zealand birds: first bird records for New Zealand 1769–2007. Raleigh, NC, Lulu Press. 278 p.
- Weins JJ 2004. What is speciation and how should we study it? American Naturalist 163: 914–923. 10.1086/386552
- Wiley EO 1978. The evolutionary species concept reconsidered. Systematic Zoology 27: 17–26. 10.2307/2412809
- Williams G 1976. The New Zealand wattlebirds (Callaeatidae). Proceedings of the Sixteenth International Ornithological Congress. Canberra, Australian Academy of Science. Pp. 161–169.
- Winker K, Rocque DA, Braile TM, Pruett CL 2007. Vainly beating the air: species-concept debates need not impede progress in science or conservation. In: Cicero C, Remsen JV eds. Festschrift for Ned K. Johnson: geographic variation and evolution in birds. Washinton, DC, The American Ornithologists' Union. Pp. 30–44.