ABSTRACT
This work extends a previous report on the outbreak of an unknown neurotoxic sea slug along the southwestern Atlantic coast by providing identification to species level. In this study a c. 550 base pair (bp) region of the cytochrome c oxidase subunit 1 mitochondrial gene used for genetic barcoding was sequenced from three individual slugs sourced from three locations in Argentina. These data, in concert with previous description on the morphology and the presence of a neurotoxin of similar effect of that produced by pleurobranchs from New Zealand, confirm the identity of these sea slugs as Pleurobranchaea maculata. This is the first record of the species in the Atlantic Ocean, thus suggesting an exotic origin. Given the fast spread of reports in the southwestern Atlantic (c. 2000 km of coastline in 6 years) we recommend urgent studies to determine the origin, toxicity, population genetics and impacts on local ecosystems.
Introduction
The ability to determine whether a newly found organism is a non-native species is critical to enable rapid management responses. Taxonomic uncertainty based solely on morphological descriptions is common among marine invertebrates, with accurate identification at species level often requiring great taxonomic expertise and the integration of evidence from diverse sources. In the last few years molecular tools have become commonplace in this integrative view of taxonomy. In particular, DNA barcoding has been demonstrated to be a powerful tool for specimen identification (Geller et al. Citation2010, 2013). DNA barcoding has been successfully used in studies dealing with the identification of marine molluscs, including gastropods (Hunt et al. Citation2010; Jennings et al. Citation2010; Jörger et al. Citation2012; Krug et al. Citation2012, Citation2013; Layton et al. Citation2014).
Recently, we reported on the sudden appearance and subsequent increase in abundance of an unknown neurotoxic sea slug of the genus Pleurobranchaea along the southwestern Atlantic coast (Farias et al. Citation2015). A few individuals were first observed in 2009 at the port of Mar del Plata (Farias Citation2009) and thereafter new records arose from several other locations in Argentina (Buenos Aires from Mar del Plata to Bahía Blanca, up to 60 m depth, and in the San Matias and Nuevo Gulfs, in the provinces of Rio Negro and Chubut respectively; NE Farias, Universidad Nacional de Mar del Plata, unpubl. data), spanning c. 2000 km of coastline.
Pleurobranchs are easily identifiable by the plumose gill located on the right side of the body. However, identification at species level is challenging, even with detailed analysis of the internal anatomy. Based solely on morphological evidence, Farias et al. (Citation2015) showed that the newly observed sea slug was not Pleurobranchaea inconspicua, the only Pleurobranchaea previously known to occur in this region (Muniain et al. Citation2006), and suggested Pleurobranchaea maculata as the most likely candidate. Individuals of P. maculata are commonly found in shallow subtidal areas around New Zealand, and have also been recorded in southeastern Australia, China, Sri Lanka and Japan (Willan Citation1983). Further evidence to support this assertion came from mouse bioassay data indicating that the sea slug contained a neurotoxin with a similar mode of action to the tetrodotoxin identified in P. maculata from New Zealand (McNabb et al. Citation2010). Therefore, obtaining molecular data to identify the species became a priority to determine whether this represented the early stages of a biological invasion. Here, we use genetic barcodes (cytochrome c oxidase subunit 1 gene [CO1]) of three individuals sourced from distant locations along the southwestern Atlantic coast to enable identification of the specimens to a species level.
Materials and methods
Sampling, DNA extraction, PCR amplification and sequencing
Single individuals were collected from three distant locations: off Buenos Aires, Argentina (38°52′57″ S 59° 35′6″ W, 40 m depth, trawling onboard the R/V Puerto Deseado-CONICET); San Antonio Oeste, Rio Negro, Argentina (40°43′37.27″ S 64°56′47.63″ W, 0 m depth, by hand); and Puerto Madryn, Chubut, Argentina (42°44′15.22″ S 65°01′40.27″ W, 3–5 m depth, SCUBA diving). Specimens were photographed, cryo-anaesthetised and fixed directly in 96% ethanol following Geiger (Citation2006). DNA was extracted from a small piece of mantle using the E.Z.N.A. Mollusc DNA Kit (Omega) according to the protocol supplied by the manufacturer. Polymerase chain reactions (PCRs) were performed in 50 µL volumes containing 25 μL of AmpliTaq Gold 360 master mix (Life Technologies), 5 μL bovine serum albumin (0.2 mg/mL; Life Technologies), 0.4 μM of each primer targeting the CO1 (dg LC01490 and dgHC02198; Geller et al. Citation2013) and template DNA (c. 20 ng). Cycling conditions were 94 °C 3 min, followed by 40 cycles of 94 °C 30 s, 50 °C 30 s, 72 °C 90 s and a final extension of 72 °C 10 min. PCR products were visualised with 1% agarose gel electrophoresis with Red Safe DNA Loading Dye and UV illumination and purified using an AxyPrep PCR cleanup kit (Axygen). Bi-directional sequencing was undertaken using the BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems).
Data analysis
The sequences of Argentinian specimens were first compared with those of P. maculata from Wood et al. (Citation2012) available on NCBI GenBank database (www.ncbi.nlm.nih.gov/GenBank) using MegaBlast (Benson et al. Citation2013), and with sequences from other Pleurobranchomorpha available in BOLD (Barcode of Life Database, http://www.boldsystems.org/) using the BOLD Identification System (Ratnasingham & Hebert Citation2007). Additionally, we assembled a CO1 barcoding dataset with all the sequences available to date of representatives of the family Pleurobranchidae from the above public databases, including the three from the Argentine specimens presented here (hereafter we will refer to this dataset simply as Pleurobranchidae). The sequences were aligned manually with BioEdit (Hall Citation1999) and the extremes cut to fit the length of the fragments obtained in this work. Sequences shorter than those of our specimens were discarded.
To determine the existence of a barcode gap in Pleurobranchidae we calculated the pairwise distance matrix for intra- and inter-specific genetic distances under the maximum composite likelihood method (Tamura et al. Citation2004). Then, we conducted an evolutionary analysis including only the sequences of the three species of the genus Pleurobranchaea, our unidentified specimens, with the aim of testing whether our sequences were clustered with those of P. maculata within the resulting phylogenetic tree. As outgroup we used a CO1 sequence of Pleurobranchus areolatus from GenBank (JN675223). The evolutionary history was inferred using the neighbor-joining method (Saitou & Nei Citation1987). The analyses were conducted using MEGA6 (Tamura et al. Citation2013)
Finally, we applied Automatic Barcoding Gap Discovery (ABGD; Puillandre et al. Citation2012) to the whole Pleurobranchidae dataset. The analysis was performed directly from the web interface (http://www.abi.snv.jussieu.fr/public/abgd/, accessed 5 January 2016). This program uses an algorithm that automatically detects significant barcode gaps in the dataset under analysis and then sorts the sequences into different partitions (hypothetical species). Under the hypothesis that our specimens were P. maculata we expected to recover the sequences from Argentina in the same partition with those P. maculata from New Zealand included in Pleurobranchidae. ABGD analyses were initially performed with the default value of relative gap width (X = 1.5) and using all the distance metrics available in the system. The other parameter values were left as defaults.
The specimens and the sequences obtained in this study were deposited in the invertebrate collection of the University of Mar del Plata and uploaded to the NCBI GenBank database, respectively, under the following voucher ID/GenBank accession numbers: UNMDP-ZINV209/KU310926; UNMDP-ZINV219/KU310927; and UNMDP-ZINV225/ KU310928.
Results and discussion
The final sequences (564 bp) obtained for the three individuals from Argentina were identical. MegaBlast analysis showed high similarity (99%) with the four sequences available for P. maculata in GenBank, all from New Zealand (GenBank accession numbers JN675220 to JN675223). Sequence similarity with other pleurobranchs in the NCBI or BOLD databases was much lower: P. meckeli from northwest Africa (MegaBlast 83%; FJ917499 and AY345026) and P. inconspicua from Florida, USA (BOLD IDS 82%, FPMAR057-08).
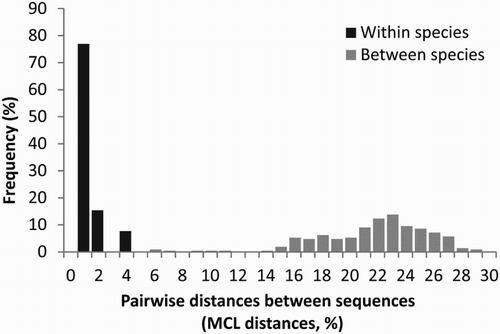
After alignment and editing, the final CO1 barcoding dataset of Pleurobranchidae consisted of 82 CO1 sequences of 530 bp length, representing 22 nominal species distributed in five genera plus our three unidentified specimens. shows that the frequency distributions of the intra-specific and inter-specific pairwise genetic distances do not overlap, thus supporting the use of CO1 barcoding for identification of representatives of Pleurobranchidae at a species level.
Figure 1. Frequency distribution of intra-specific and inter-specific pairwise genetic distances in Pleurobranchidae. The plot is based on 82 sequences of 530 bp obtained from BOLD system representing 22 nominal species distributed in five genera plus the three specimens from Argentina presented in this work. Pairwise genetic distances were calculated under the maximum composite likelihood (MCL) method.
The neighbor-joining analysis produced a tree with three groups in accordance to the species assigned a priori. The three specimens from Argentina were clustered with the specimens of P. maculata from New Zealand, as predicted ().
Figure 2. Evolutionary relationship of the genus Pleurobranchaea. Neighbor-joining phylogenetic tree performed with 15 CO1 sequences of 530 bp from representatives of the genus Pleurobranchaea. The optimal tree with the sum of branch length = 0.272 is shown. Numbers above nodes are bootstrap support values (1000 replicates).The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. Terminal values are the identifiers for the individual CO1 sequences available for Pleurobranchaea maculata in GenBank and of P. meckeli and P. inconspicua in the Public Data Portal of the BOLD system.
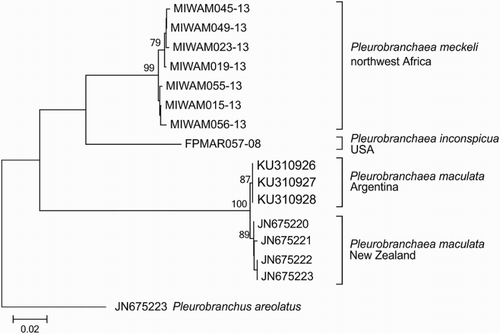
During the ABGD analysis the relative width of barcoding gap (X-value) had to be lowered to 1 to get any partitions, the rest of the parameters were left as default. In full agreement with our a priori delimitation of species, the ABGD grouped the sequences into 22 partitions (hypothetical species) with a prior of intra-specific divergence up to 0.0129 irrespective of the model used to calculate genetic distances. The three specimens from Argentina were always recovered in the same group with the four sequences of P. maculata from New Zealand.
The congruence of the results from the molecular evidence analysed by independent methods, in concert with the morphological descriptions in Farias et al. (Citation2015), confirms the identification of the recently observed sea slug along the southwestern Atlantic coast as Pleurobranchaea maculata. Given the presence of neurotoxins in Argentinian specimens (Farias et al. Citation2015), and the close CO1 sequence similarity, it is possible that New Zealand populations may be the original source for this incursion. Further genetic studies with more individuals from each locality and different populations from the putative native distribution are required to test this hypothesis.
Pleurobranchaea maculata has many of the traits described for successful invaders (Whitney & Gabler Citation2008; Sargent & Lodge Citation2014): fast generation times (Gibson Citation2003; Khor et al. Citation2014); high likelihood of reproduction at very low population densities since they are hermaphrodite (Puurtinen & Kaitala Citation2002); planktotrophic larvae with long development in the water column which favours wide dispersion (Gibson Citation2003); the ability to survive at extremely high densities (e.g. up to 0.8 individuals m−2, Khor et al. Citation2014; Taylor et al. Citation2015); and a diet with a wide range of (new) food types (SA Wood, Cawthron Institute, unpubl. data; Ottaway Citation1977, under the synonym P. novaezelandiae; Willan Citation1984). Given these life history traits, the extensive geographical spread, the absence of records in previous studies within the region (Carcelles Citation1944; Carcelles & Williamson Citation1951; Bastida Citation1971; Valentinuzzi de Santos Citation1971; Scarabino Citation1977; Bremec Citation1989; Olivier et al. Citation1966a,b; Genzano et al. Citation2011) and the rapid increase in local abundances (NE Farias, S Obenat and E Schwindt, pers. obs.), it is possible that this represents a new incursion of an invasive species. Further studies on P. maculata’s toxicity, population genetics and impact on local ecosystems are urgently needed.
Acknowledgements
The authors thank Marcos Tatián (IDEA-UNC-CONICET), Fabrizio Scarabino (National Museum of Natural History of Uruguay) and Henrique Kanck de Almeida (UBA-CONICET) for their collaboration in the collection of samples, and Laura Biessy (Cawthron) for technical assistance.
Associate Editor: Dr Jonathan Banks.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Bastida RO. 1971. Las incrustaciones biológicas en el puerto de Mar del Plata. Período 1966/67. Revista Del Museo Argentino de Ciencias Naturales ‘Bernardino Rivadavia. 3:203–285.
- Benson DA, Cavanaugh M, Clark K, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW. 2013. GenBank. Nucleic Acids Research. 41:D36–D42. doi: 10.1093/nar/gks1195
- Bremec CS. 1989. Macrobentos del área de Bahía Blanca (Argentina). Inventario faunístico. Investigaciones Marinas (La Paz, México). 4:175–190.
- Carcelles AR. 1944. Catálogo de los moluscos marinos de Puerto Quequén (República Argentina). Revista Del Museo de La Plata (Argentina) (NS, Zoologia). 3:233–309.
- Carcelles AR, Williamson S. 1951. Catálogo de los moluscos marinos de la Provincia Magallánic. Revista Del Museo Argentino de Ciencias Naturales (Zoologia). 2(5):225–383.
- Farias NE. 2009. (Nov 23) Pleurobranchaea from Argentina. [Message in] Sea Slug Forum. Australian Museum, Sydney. Available from: http://www.seaslugforum.net/find/22819.
- Farias NE, Obenat S, Goya ABA. 2015. Outbreak of a neurotoxic side-gilled sea slug (Pleurobranchaea sp.) in Argentinian coasts. New Zeal J Zool. 42:51–56. doi: 10.1080/03014223.2014.990045
- Geiger DL. 2006. Marine gastropoda. In: Sturm CF, Pearce TA, Valdés Á, editors. The Mollusks: a guide to their study, collection, and preservation. Boca Ratón, FL: Universal Publishers; p. 295–312.
- Geller JB, Darling JA, Carlton JT. 2010. Genetic perspectives on marine biological invasions. Ann Rev Mar Sci. 2(1):367–393. doi: 10.1146/annurev.marine.010908.163745
- Geller J, Meyer C, Parker M, Hawk H. 2013. Redesign of PCR primers for mitochondrial cytochrome c oxidase subunit I for marine invertebrates and application in all-taxa biotic surveys. Mol Ecol Resour. 13(5):851–861. doi: 10.1111/1755-0998.12138
- Genzano G, Giberto D, Bremec C. 2011. Benthic survey of natural and artificial reefs off Mar del Plata, Argentina, southwestern Atlantic. Lat Am J Aquat Res. 39(3):553–566. doi: 10.3856/vol39-issue3-fulltext-15
- Gibson GD. 2003. Larval development and metamorphosis in Pleurobranchaea maculata, with a review of development in the notaspidea (Opisthobranchia). Biol Bull. 205:121–32. doi: 10.2307/1543233
- Hall TA. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl. Acids. Symp. Ser. 41:95–98.
- Hunt B, Strugnell J, Bednarsek N, Linse K, Nelson RJ, Pakhomov E, Würzberg L. 2010. Poles apart: the “Bipolar” pteropod species Limacina helicina is genetically distinct between the Arctic and Antarctic Oceans. PLoS ONE. 5(3):e9835. doi: 10.1371/journal.pone.0009835
- Jennings RM, Bucklin A, Ossenbrügger H, Hopcroft RR. 2010. Species diversity of planktonic gastropods (Pteropoda and Heteropoda) from six ocean regions based on DNA barcode analysis. Deep-Sea Res Pt II. 57:2199–2210. doi: 10.1016/j.dsr2.2010.09.022
- Jörger KM, Norenburg JL, Wilson NG, Schrödl M. 2012. Barcoding against a paradox? Combined molecular species delineations reveal multiple cryptic lineages in elusive meiofaunal sea slugs. BMC Evol Biol. 12(1):245. doi: 10.1186/1471-2148-12-245
- Khor S, Wood S, Salvitti L, Taylor DI, Adamson J, McNabb P, Cary SC. 2014. Investigating diet as the source of tetrodotoxin in Pleurobranchaea maculata. Mar Drugs. 12:1–16. doi: 10.3390/md12010001
- Krug PJ, Asif JH, Baeza I, Morley MS, Blom WM, Gosliner TM. 2012. Molecular identification of two species of the carnivorous sea slug Philine, invaders of the US west coast. Biol Invasions. 14(11):2447–2459. doi: 10.1007/s10530-012-0242-9
- Krug PJ, Vendetti JE, Rodriguez AK, Retana JN, Hirano YM, Trowbridge CD. 2013. Integrative species delimitation in photosynthetic sea slugs reveals twenty candidate species in three nominal taxa studied for drug discovery, plastid symbiosis or biological control. Mol Phylogenet Evol. 69(3):1101–1119. doi: 10.1016/j.ympev.2013.07.009
- Layton KKS, Martel AL, Hebert PDN. 2014. Patterns of DNA barcode variation in Canadian marine molluscs. PLoS ONE. 9(4):e95003. doi: 10.1371/journal.pone.0095003
- McNabb P, Selwood AI, Munday R, Wood SA, Taylor DI, Mackenzie LA, van Ginkel R, Rhodes L, Cornelisen C, Heasman K, et al. 2010. Detection of tetrodotoxin from the grey side-gilled sea slug—Pleurobranchaea maculata, and associated dog neurotoxicosis on beaches adjacent to the Hauraki Gulf, Auckland, New Zealand. Toxicon: Official Journal of the International Society on Toxinology. 56:466–473. doi: 10.1016/j.toxicon.2010.04.017
- Muniain C, Ardila NE, Cervera JL. 2006. Pleurobranchaea inconspicua Bergh, 1897 (Opisthobranchia: Pleurobranchidae): redescription and distribution from Argentina and Colombia. Bonner Zoologische Beiträge. 55:291–300.
- Olivier SR, Escofet A, Orensanz JML, Pezzani SE, Turró AM, Turró ME. 1966a. Contribución al conocimiento de las comunidades bentónicas de Mar del Plata. I. Las costas rocosas entre Playa Grande y Playa Chic. Anales de La Comisión de Investigaciones Científicas de La Provincia de Buenos Aires (Argentina). 7:185–206.
- Olivier SR, Kreibhom de Paternoster I, Bastida RO. 1966b. Estudios biocenóticos en las costas de Chubut (Argentina) I. Zonación biocenológica de Puerto Pardelas (Golfo Nuevo). Instituto de Biología Marina (Mar Del Plata, Argentina), Boletín. 10:1–74.
- Ottaway J. 1977. Pleurobranchaea novaezelandiae preying on Actinia tenebrosa. New Zeal J Mar Fresh. 11:125–130. doi: 10.1080/00288330.1977.9515665
- Puillandre N, Lambert A, Brouillet S, Achaz G. 2012. ABGD, automatic barcode gap discovery for primary species delimitation. Mol Ecol. 21(8):1864–1877. doi: 10.1111/j.1365-294X.2011.05239.x
- Puurtinen M, Kaitala V. 2002. Mate-search efficiency can determine the evolution of separate sexes and the stability of hermaphroditism in animals. Am Naturalist. 160(5):645–660. doi: 10.1086/342821
- Ratnasingham S, Hebert PDN. 2007. BARCODING BOLD: the barcode of life data system (http://www.barcodinglife.org). Mol Ecol Notes. 7:355–364.
- Saitou N, Nei M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 4:406–425.
- Sargent LW, Lodge DM. 2014. Evolution of invasive traits in nonindigenous species: increased survival and faster growth in invasive populations of rusty crayfish (Orconectes rusticus). Evol Appl. 7(8):949–961. doi: 10.1111/eva.12198
- Scarabino V. 1977. Moluscos del Golfo San Matías (Prov. De Río Negro, Rep. Argentina). Comunicaciones de La Sociedad Malacológica Del Uruguay. 4:177–286.
- Tamura K, Nei M, Kumar S. 2004. Prospects for inferring very large phylogenies by using the neighbor-joining method. P Natl A Sci (USA). 101:11030–11035. doi: 10.1073/pnas.0404206101
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30:2725–2729. doi: 10.1093/molbev/mst197
- Taylor D, Wood S, McNabb P, Ogilvie S, Cornelisen C, Walker J, Khor S, Cary S. 2015. Facilitation effects of invasive and farmed bivalves on native populations of the sea slug Pleurobranchaea maculata. Mar Ecol Prog Ser. 537:39–48. doi: 10.3354/meps11466
- Valentinuzzi de Santos S. 1971. Estudio preliminar sobre las comunidades intercotidales del Puerto Ing. White. Physis (Argentina). 30(81):407–416.
- Whitney KD, Gabler CA. 2008. Rapid evolution in introduced species, ‘invasive traits’ and recipient communities: challenges for predicting invasive potential. Diver Distrib. 14(4):569–580. doi: 10.1111/j.1472-4642.2008.00473.x
- Willan RC. 1983. New Zealand side-gilled sea slugs (Opistobranchia: Notaspidea: Pleurobranchidae). Malacologia. 23(2):221–270.
- Willan RC. 1984. A review of diets in the Notaspidea (Mollusca: Opisthobranchia). J Malacol Soc Aust. 6:125–142.
- Wood SA, Taylor DI, McNabb P, Walker J, Adamson J, Cary SC. 2012. Tetrodotoxin concentrations in Pleurobranchaea maculata: temporal, spatial and individual variability from New Zealand populations. Mar Drugs. 10:163–176. doi: 10.3390/md10010163
