ABSTRACT
The Melolonthinae branch of the beetle family Scarabaeidae has evolved in isolation in Aotearoa, radiating into >100 endemic species, since Aotearoa separated from Gondwanaland 82 million years ago. The group includes important pasture pests, such as the New Zealand grass grub Costelytra giveni and the manuka beetle Pyronota festiva. These beetles, like other organisms, host their own distinctive microflora including beneficial microbial symbionts and pathogens. A wide range of microbial pathogens infect the Scarabaeidae, but in Aotearoa the bacteria Serratia entomophila, S. proteamaculans and S. quinivorans (Enterobacteriaceae) are frequently found causing natural disease epizootics in C. giveni. S. entomophila is widespread in Aotearoa pasture soils, with only rare isolations of S. entomophila documented in other countries. In contrast S. proteamaculans and S. quinivorans are globally ubiquitous, and are widely distributed within Aotearoa, with some isolates active against either C. giveni or Pyronota spp. larvae, or both. Virulence determinants that impart differential host specificity and potency are located on variants of the amber disease associated plasmid (pADAP). The host specificity of the Serratia-scarab system and the absence of similar systems in other geographies, suggests that the relationship between Serratia spp. and endemic scarabs has evolved in Aotearoa.
Introduction
The ancestors of the beetle family Scarabaeidae (Coleoptera) can be traced to the Mesozoic era, more than 200 million years ago, and followed an evolutionary radiation across the developing continents (Morón Citation1984). Scarab beetles, of which there are more than 30,000 species belonging to 1600 genera (Grebennikov et al. Citation2004), are found in a broad range of ecosystems, from tropical to temperate climates, with many being recognised as destructive grassland pests (Klein et al. Citation2007). The Gondwanaland fragment which later became Aotearoa separated some 82 million years ago (Rey and Muller Citation2010) together with the precursors of melolonthine scarabs which have radiated into more than 120 endemic species in Aotearoa (Given and Hoy Citation1952). Despite their wide diversity, scarab beetles have several properties in common. Larvae develop through three larval instars feeding on decaying plant material, dung or roots in the soil. The larval gut is adapted to this diet with a large fermentation chamber for microbial degradation of the ingested material (Zhang and Jackson Citation2008). The larvae make chambers and pupate in the soil or other concealed locations before emerging as adults, often synchronously in swarms, for feeding on leaves and fruit, mating, and dispersal. The Aotearoa melolonthinae scarabs appear to have evolved in the native grasslands and bush margins with availability of food for both larval and adult stages, but most remain rare in the native habitat. However, some species, such as the New Zealand grass grub (Costelytra giveni) (Coca-Abia and Romero-Samper Citation2016) (formerly C. zealandica), known as tūtae ruru by Māori, and the mānuka beetles (Pyronota spp.), known as kēkerewai or repowai by Māori, have adapted to feed on introduced pasture species in the modified Aotearoa landscape (LeFort et al. Citation2014) and have become major agricultural pests (Ferguson et al. Citation2019). High densities of C. giveni larvae decimate the pasture root systems leading to browning of grasses typically observed mid to late March. This severing of the roots enables the ‘lifting’ of the sward layer when pulled (). C. giveni is widely dispersed throughout Aotearoa, but has been absent from the West Coast, South Island (Jackson et al. Citation1991). However, in 2012 a localised population, likely accidently introduced, was identified near the Westport airport (Townsend et al. Citation2013), and this population has now spread to Cape Foulwind, where it has become established (Hurst et al. Citation2022).
Figure 1. Damage caused by grass grub. A, Grass grub damage manifesting as dead patches in pasture (Canterbury). B, uplifted sward revealing high C. giveni larval densities with larvae forming chambers as depicted in C.

Mānuka beetle larvae cause similar root damage as C. giveni larvae and typically inhabit areas with light soils (Thomson et al. Citation1979). Although Pyronota spp. (P. festiva, P. laeta, and P. setosa) are smaller in size than C. giveni, the larvae of both are similar in appearance (). P. festiva is widely distributed throughout Aotearoa, while P. setosa has a scattered, mainly littoral, distribution in sandy soils in coastal scrubland areas (Brown Citation1963). Pasture damage attributed to Pyronota species has mainly been reported on light soils in proximity to native vegetation or in areas of pasture regrowth, typified by large scale pasture renovation (Thomson et al. Citation1979; Jackson et al. Citation2012; Townsend et al. Citation2018). P. setosa has caused extensive pasture damage following large scale pasture renovation at Cape Foulwind, West Coast, South Island (Dunbar et al. Citation2012). Pyronota spp. and C. giveni often coexist in pastures near bushland or after pasture renovation in hill country.
Figure 2. Photographs of grass grub and mānuka beetle larvae. A, C. giveni. B, P. festiva. Scale bar 5 mm.

Both C. giveni and Pyronota spp. share a similar univoltine seasonal lifestyle, with their eggs typically laid in November–December (late Spring) with feeding damage by the larvae most apparent from autumn (late March–April) to winter (August). The larvae go through three larval instars prior to pupating late August to emerge in November as adults (Thompson et al. Citation1979). Under adverse climatic conditions such as drought, grass grub larvae can alter their metabolism and enter a two-year life cycle.
In the primary sector these pest species mainly affect, but are not restricted to, pasture comprising ryegrass or clover. Annual pasture losses due to grass grub beetle were estimated at NZ$140–380M for dairy and $75–205M for sheep and beef (Ferguson et al. Citation2019). This excludes the arable sector comprising an area >160,000 ha, or additional niche cropping markets such vineyards, where adult C. giveni feed on foliage (A,B). In establishing pastures, C. giveni larvae typically reach damaging levels 2–4 years after sowing (Jackson et al. Citation2017). In some pastures populations of greater than 500 larvae/m2 are common and over 1000 larvae/m2 have sometimes been recorded (Jackson et al. Citation2012). C. giveni larval numbers greater than 150/m2 are considered damaging to pasture (Ferguson et al. Citation2019). Due to their smaller size, mānuka beetle can exceed larval densities of 2000/m2 of pasture, although populations greater than 300 larvae/m2 significantly impact pasture performance (Dunbar et al. Citation2012).
Pathogens of endemic scarabs
The evolution of scarabs in microbial-rich soil appears to have led to a high degree of resistance to generalist pathogens and the development of a wide range of scarab-specific pathogens including fungi, viruses, rickettsia, bacteria, and protozoa. The scarabs in Aotearoa are hosts to a particular microbiota including both symbionts (Zhang and Jackson Citation2008) and pathogens (Glare, O’Callaghan, et al. Citation1993). Scarabaeid beetles in general are known to be susceptible to a wide range of pathogens from all classes and some of these microorganisms can play an important role in population regulation. Pathogens are particularly abundant, with more than 30 species of microbial pathogens or entomopathogenic nematodes recorded from C. giveni alone (Bourner et al. Citation1996). The pathogens of Aotearoa Scarabaeidae include obligate pathogens such as the bacterium Paenibacillus popilliae (Klein and Jackson Citation1992) which is ubiquitous, but specific for the Scarabaeidae. Strains of P. popilliae are found in scarab populations from all continents, and many are host specific, which suggests that the pathogens arrived and evolved with the founder populations. Species of entomopathogenic fungi attacking Scarabaeidae found in Aotearoa, such as some Metarhizium spp., have a wider host range with susceptible hosts across a range of insect orders (Glare et al. Citation2021). However, even among the ‘broad spectrum’ fungi, strain specificity and efficacy can differ (Brunner-Mendoza et al. Citation2019). Metarhizium novozealandicum is the most common fungal pathogen of grass grub in Aotearoa (Villamizar et al. Citation2021).
Notably the Aotearoa scarabs are also hosts to a range of facultative bacterial pathogens which can live and reproduce on alternative substrates outside the host. These include non-sporeforming bacteria from the Enterobacteriaceae (Serratia spp.) which are found to cause epizootics among Aotearoa scarabs. These bacteria can be found in the soil in both pathogenic and non-pathogenic forms (Grimont et al. Citation1988) and are abundant in long established scarab beetle populations. A less prevalent pathogen is the broad-spectrum entomopathogen Yersinia entomophaga, isolated from C. giveni larvae, which is highly active towards scarab species, but has yet to be reisolated from a scarab collected from the field (Hurst, Becher, Young, et al. Citation2011a).
Serratia spp. as pathogens of grass grub
In 1981, a honey/amber colouration was recognised as a symptom of ill health in C. giveni larvae and found to be caused by a bacterial infection (Trought et al. Citation1982). The disease, initially termed ‘honey disease’, was recognised as being widespread and the cause of population decline (Jackson Citation1984) with disease symptoms caused by two species of soil bacteria (Stucki et al. Citation1984). Due to concern by beekeepers about the name ‘honey disease’, the disease was later designated amber disease, based on the distinctive colouration of the infected larvae. Grimont et al. (Citation1988) classified the pathogenic isolates into two species, Serratia entomophila sp. nov. and Serratia proteamaculans, based on definitive phenotypic and substrate utilisation tests. The newly defined S. entomophila was originally represented by nineteen isolates from diseased C. giveni larvae collected in Aotearoa and a single isolate designated 222, from the Institute Pasteur, France. S. entomophila can be differentiated from other Serratia spp. through the ability to utilise itaconate and further differentiated into two biotypes based on further biochemical tests (Grimont et al. Citation1988).
Since the initial study, S. entomophila has been frequently isolated from C. giveni larvae and pasture soils in Aotearoa (O’Callaghan et al. Citation1999) but, to date, despite extensive searches only two further isolates of S. entomophila have been reported from outside Aotearoa. These include the Mexican isolate Mor.4.1, reported to control the scarab Phyllophaga blanchardi (Nunez-Valdez et al. Citation2008), and S. entomophila strain AB2, isolated from Heliothis armigera larvae in India (Chattopadhyay et al. Citation2011), which has shown pathogenic activity against a range of lepidopteran larvae (Chattopadhyay et al. Citation2012).
In contrast, S. proteamaculans is a ubiquitous bacterium (Grimont and Grimont Citation2006) which, based on 16S ribosomal RNA phylogeny, is placed within the Serratia liquefaciens-proteamaculans-grimesii-quinivorans complex (Begrem et al. Citation2021). Bacteria of this group have been readily isolated from the phytosphere (Grimont et al. Citation1981). Subsequently Serratia proteamaculans subsp. quinovora was reclassified as S. quinivorans using DNA–DNA cross hybridisation and substrate utilisation profiling (Ashelford et al. Citation1983). Using genomic comparisons based on Average Nucleotide Identity (ANI), Williams et al. (Citation2022) recently found that S. proteamaculans and S. quinivorans formed separate phylogroups. As detailed below, Aotearoa scarab pathogens have been found belonging to both of these phylogroups.
Using a wide range of historical isolates and genomic approaches, Williams et al. (Citation2022) confirmed the phylogeny of S. entomophila which was only found among the Aotearoa isolates and the single French isolate 222. Genomic analysis by these authors defined three lineages within S. entomophila; lineage 17 included isolates from the offshore Chatham Islands, and lineage 18 comprised isolates from a single site in Canterbury. The remaining lineage 19 included isolates collected from around Aotearoa and the French isolate 222. Lineages 17 and 18 correspond to Grimont biotype 1 while lineage 19 corresponds to Grimont biotype 2.
In Aotearoa both pathogenic and non-pathogenic isolates of S. entomophila and S. proteamaculans (some now reclassified as S. quinivorans) have been isolated from insects and soil, with the pathogenic forms more likely to be associated with the presence of the host insects (Glare, Corbett, et al. Citation1993; Dodd et al. Citation2006). Pathogenicity is defined by bioassay through development of symptoms after ingestion of bacteria (Jackson and Saville Citation2000). Surprisingly, host range testing found pathogenic S. entomophila are specific in causing amber disease in C. giveni but had no effect on other scarabs or other insect species (Jackson et al. Citation1991; Jackson et al. Citation1998; Jackson Citation2003). More recently, and as later outlined, isolates of S. proteamaculans and S. quinivorans have been identified with dual activity towards Pyronota spp. and C. giveni (Hurst et al. Citation2018; Hurst et al. Citation2021).
Amber disease – a chronic infection
An outline of the amber disease process in presented in . After ingestion of a pathogenic dose of an amber disease-causing isolate of S. entomophila or S. proteamaculans, C. giveni larvae rapidly cease feeding and the larval gut, which normally contains dark digestive fluid, clears and the larvae take on a characteristic amber colouration (F) before entering a long quiescent phase and finally death through septicaemia (Jackson et al. Citation1993; Jackson et al. Citation2001). There is no lethal or sub-lethal effect on the larvae from ingestion of non-pathogenic strains, even at very high doses. Ingested bacteria are mostly flushed into the large fermentation chamber prior to cessation of feeding, with some colonisation of cuticular surfaces of the gut and mouthparts (Jackson et al. Citation2001). Following gut clearance, bacterial growth occurs principally in the foregut, including the oral cavity, and hindgut of the infected larva (Jackson et al. Citation2001) and reaches more than 1 × 107 bacteria per larva in the chronic phase of infection. Gut clearance is associated with a sharp decrease in levels of the major gut digestive enzymes, trypsin and chymotrypsin (Gatehouse et al. Citation2009; Jackson Citation1995), associated with inhibition of exocytosis in the midgut cells (Gatehouse et al. Citation2008). As the disease progresses the infected larva becomes moribund, fat cells atrophy, and the body develops the clear amber appearance characteristic of the disease (Jackson et al. Citation1993). In contrast to infections by toxin-bearing Bacillus thuringiensis in other insects, there is no destruction or blebbing of the midgut epithelium and the infective bacteria remain within the larval gut until the final stages of infection (Jackson et al. Citation1993; Jackson et al. Citation2001). In the infected larva, bacteria can be visualised attached to food particles () or gut membranes but no specific sites of S. entomophila colonisation associated with infection have been identified (Wilson et al. Citation1992; Jackson et al. Citation2001; Hurst and Jackson Citation2002). As time progresses the infected larva becomes weaker before bacteria eventually invade the haemocoel through the weakened membranes to cause death of the insect by septicaemia and further replication of the bacteria on the cadaver (Jackson et al. Citation2001). The amber disease process can take more than 4 months from infection to death of larvae and is defined as a chronic infection.
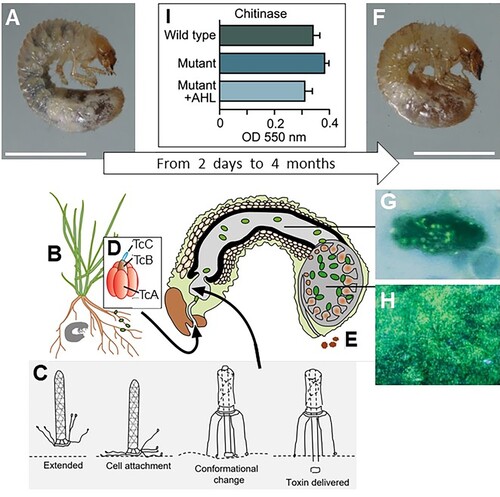
Figure 4. Overview of the S. entomophila – C. giveni system. (A), Healthy C. giveni larvae (Scale bar 5 mm) ingest the roots (B) and associated S. entomophila. Within 2–5 days the larvae cease feeding, a result of as few as 500 anti-feeding prophage (Afp) particles. The proposed model for the action of the Afp is depicted in (C), where the extended virus-like Afp particle latches onto a currently undetermined receptor on cuticular membranes or the intestinal cells using its tail fibres. Once bound (cell attachment), the Afp particle changes conformation to inject the Afp loaded toxin into the eukaryotic target cell. The S. entomophila pathogenicity (Sep) Toxin Complex (Tc) (D) is released by ingested bacteria. This causes gut clearance, expulsion of gut contents (E) and the larvae to become amber in colour (F); scale bar 5 mm. The disease process can last for more than 4 months. There are no apparent sites of S. entomophila colonisation on the surface of midgut cells, with bacteria only adhering to particulate matter of the gut and membranous surfaces (G). Within 6 days of ingestion, bacteria multiply within the fermentation chamber of the insect (H). Under conditions of high cell density levels of the S. entomophila-derived chitinases are reduced (I), possibly prolonging the time of infection. I, measurements of chitinase activity; WT, denotes wildtype S entomophila; Mutant, denotes quorum sensing (QS) mutant; +AHL denotes the mutant complemented by addition of an exogenous supply of the QS signal molecule N-Acylhomoserine lactone (AHL). Figure modified from Hurst (Citation2016).
Defining the genetic determinants of amber disease
Ecological studies showed that both S. entomophila and S. proteamaculans could be isolated from diseased C. giveni larvae, but both species occurred in pathogenic and non-pathogenic forms (Grimont et al. Citation1988, O’Callaghan et al. Citation1988). This suggested that a transferable genetic element was involved in amber disease. Through plasmid visualisation, Glare, Corbett, et al. (Citation1993) found that amber disease-causing isolates of S. entomophila and S. proteamaculans both contained a similar 153 kb plasmid that was absent in non-pathogenic isolates. Removal of the plasmid through heat curing resulted in a total loss of virulence. The amber disease encoding plasmid was then designated pADAP for amber disease associated plasmid. Assessment of the DNA restriction enzyme profiles of both the plasmids and genomes of 21 virulent S. entomophila isolates showed that they were all quite similar, with only a single S. entomophila pADAP variant (p626) identified which differed by a single RE-DraI 4.2-kb band. In contrast DraI plasmid profiles of S. proteamaculans strains revealed a wide range of pADAP variants (Dodd et al. Citation2006).
Variation in pADAP was later examined through the genomic sequencing of plasmids from 51 S. entomophila and S. proteamaculans isolates derived from diverse geographies and exhibiting diverse disease phenologies (Sitter et al. Citation2021). Ten sequenced S. entomophila pADAP plasmids showed high homology sharing >99% nucleotide identity. In contrast, the 41 sequenced S. proteamaculans pADAP plasmids were highly heterogenous in the accessory genes that they encoded despite sharing a common ∼63 kb plasmid backbone (Sitter et al. Citation2021). This study identified and characterised 12 pADAP variants designated A-L. These 12 variants differed only in their accessory regions, several of which encoded different toxin complex (Tc) variants, and these pADAP variants were termed STAMPs for Serratia transmissible adaptive mega-plasmids. Two of these STAMP variants reside in non pathogenic Serratia liquefaciens isolates 376 and 377 (Sitter et al. Citation2021).
Using a modified pADAP variant, Grkovic et al. (Citation1995) were able to conjugate pADAP into other members of the Enterobacteriaceae, including strains of Serratia marcescens, S. liquefaciens, Enterobacter agglomerans, a Klebsiella species and Escherichia coli. In all cases, receipt of the plasmid resulted in bacteria that caused amber disease symptoms in the grass grub host. Only pathogenic variants of pADAP have been found in strains of S. entomophila and S. proteamaculans in the field (Jackson et al. Citation1991), suggesting that these species encode factors that favour retention of the plasmid.
Through a process of targeted mutagenesis and DNA sequence analysis, the 153 kb pADAP was found to encode two virulence-associated regions. The first region is the sepABC (S. entomophila pathogenicity) (Hurst et al. Citation2000) gene cluster encoding a C. giveni active Tc (toxin complex) (D) (Hurst et al. Citation2000). Tcs were first identified in the chromosome of the bacterium Photorhabdus luminescens (Bowen et al. Citation1998) and have since been identified in chromosomes of Pseudomonas spp. (Rangel et al. Citation2016), Xenorhabdus nematophilus (Morgan et al. Citation2001), members of Yersinia spp. (Fuchs et al. Citation2008, Hurst et al. Citation2011a), and B. thuringiensis (Blackburn et al. Citation2011), and other bacteria (Song et al. Citation2021). Typically, Tcs are chromosomally encoded to produce three proteins, designated TcA, TcB, and TcC (D), which combine to form the final insect-active complex (ffrench-Constant and Waterfield Citation2005). The TcA component enables the delivery of the toxin effector and imparts host range specificity (Sergeant et al. Citation2006, Meusch et al. Citation2014). The TcC component is a two-domain protein comprising a distinct C-terminal effector domain (Hurst et al. Citation2000) and a conserved Rhs N-terminal domain which, together with the TcB component, envelopes the TcC C-terminal effector (Busby et al. Citation2013), allowing its docking to the TcA delivery component. Once in the midgut the Sep protein causes flushing of the gut and prevents the secretion of digestive enzymes such as trypsin and chymotrypsin by interfering with exocytosis leading to symptoms of amber disease (Gatehouse et al. Citation2009). Of interest C. giveni larvae challenged with low amounts of purified Sep proteins cleared the gut and turned amber within 5 days but later reverted to the healthy phenotype (Hurst, Jones, et al. Citation2007). This reversion did not occur after antibiotic elimination of the bacteria (Jackson et al. Citation2001).
The second pADAP virulence determinant is a pyocin-like particle called the anti-feeding prophage (Afp) (Hurst et al. Citation2004; Desfosses et al. Citation2019), variants of which have been identified in a diverse range of microbes (Sarris et al. Citation2014; Chen et al. Citation2019) and have been collectively termed extracellular contractile injection machines (eCIS) (Böck et al. Citation2017). Following ingestion by C. giveni larvae, as depicted in C, the Afp is proposed to bind via its tail fibres to a yet-to-be-defined target cell, from where it then contracts to deliver its protein effector payload to the cell cytosol, resulting in cessation of feeding activity (Hurst et al. Citation2004; Hurst, Beard, et al. Citation2007). The Afp induces an effect independently of the bacterial cell; Rybakova et al. (Citation2013) found that as few as 500 Afp particles were required to elicit cessation of feeding activity in 50% of the larval population three days post per os challenge. Higher concentrations of Afp caused a rupture of the gut membranes inducing a brown bacterial septicaemia and death within days of ingestion (Hurst et al. Citation2004). In addition to these findings, unlike other Serratia species, a targeted mutation of the S. entomophila cell density-dependent quorum sensing (QS) gene, exhibited increased chitinase production under in vitro conditions (I; Hurst Citation2016), a finding that may have implications for amber disease progression as discussed further below.
The disease process in C. giveni caused by S. entomophila shows some similarities to that of the obligate Gram-positive pathogen Paenibacillus popilliae (Bacilliae) (Klein and Jackson Citation1992). Both pathogens grow within their hosts without triggering any apparent host responses, such as melanisation or recruitment of haemocytes. P. popilliae invades midgut cells and penetrates the haemocoel where it multiplies in the haemolymph without reaction. In contrast, S. entomophila colonises and multiplies in the insect intestine. In both systems events are triggered that lead to cessation of feeding and the depletion of the larval fat bodies, leading to starvation and death of the host. In both scenarios it appears that the purpose of pathogenicity is to provide an environment suitable for replication of the pathogen.
Atypical disease-causing Serratia isolates
While both S. entomophila and S. proteamaculans pathogenic isolates generally produce similar symptoms of amber disease in C. giveni larvae, several Serratia isolates have been identified that cause atypical disease symptoms. These include isolate S. proteamaculans 143 (now S. quinivorans 143) which caused only 60–70% of challenged C. giveni larvae to exhibit an amber colour (Glare, Corbett, et al. Citation1993; Hurst, Becher, O’Callaghan Citation2011b) regardless of the dose of bacteria administered. Recently, several S. proteamaculans and S. quinivorans isolates recovered from either diseased C. giveni or Pyronota spp. larvae, sourced from a range of geographies, were found to cause heightened virulence and/or affected the larvae of both C. giveni and Pyronota spp. Unlike the stark cessation of feeding activity caused by S. entomophila, many of the larvae challenged with these S. proteamaculans or S. quinivorans isolates continued to feed but at a reduced capacity. The diseased larvae cleared their guts, transitioning to an amber phenotype and then turned brown prior to their death within 5–12 days of pathogen ingestion. In some cases, these isolates did not cause symptoms in all challenged larvae, with only 80–90% of the challenged larvae symptomatic and the remaining larvae appeared healthy over the duration of the 12-day bioassay (Hurst et al. Citation2021).
Bioinformatic analysis of the genomes of the atypical S. proteamaculans/S. quinivorans isolates and S. entomophila revealed the presence of seven distinct C. giveni or Pyronota spp. active Tcs and an Afp variant (). These included four distinct tcA (tcA1–tcA4) and tcC (tcC1–tcC4) variants with three of the tc clusters encoding two different TcC proteins. Several of the S. proteamaculans and S. quinivorans Tc-encoding isolates had activity towards both Pyronota spp. and C. giveni, with other isolates being specific to either Pyronota spp. or C. giveni (). Two S. quinivorans isolates, C and Man4, were found to cause an incomplete disease state, with the challenged larvae reverting from diseased to healthy (Hurst et al. Citation2021), a reversion effect like that previously observed with C. giveni larvae challenged with low levels of Tc Sep proteins. Differences in both host range (isolate Cfb affects both C. giveni and Pyronota spp.; ) and virulence capacity (e.g. C. giveni-specific isolate 143; ) of isolates encoding the same virulence determinants were also noted. Other isolates, such as S. proteamaculans 28F and S. quinivorans MH5, exhibited variable pathogenicity over different bioassays, being effective against C. giveni larvae one season but not the next. Although yet to be proven, this seasonal variability may reflect genetic differences in the larvae sourced from different geographic locations used for bioassay assessments, as noted by Richards et al. (Citation1997). The S. entomophila sep Tc-encoding region is phylogenetically distinct to the more closely related S. proteamaculans and S. quinivorans Tc orthologues (Hurst et al. Citation2021). When the highly variable TcC effector domain was excluded from the analysis, the divergence in these Tc clusters was more pronounced across the various Serratia TcA components which, based on studies of P. luminescens Tcs (Gatsogiannis et al. Citation2013; Meusch et al. Citation2014), impart host specificity.
Figure 5. Schematic of the S. entomophila (Se), S. proteamaculans (Sp) and S. quinivorans (Sq) Afp and Tc variants and their associated C. giveni (Cg), P. festiva (Pf), and P. setosa (Ps) host. Patterned arrows de note the Tc (TcA1-A4, TcC1 – C4) variants. Relevant isolate identifiers are listed where bold text denotes the previously documented S. entomophila Sep and Afp and the S. quinivorans isolate 143, and AGR96X AfpX virulence-associated region. Isolate names are listed where superscript letters indicate: ano activity against Pyronota species, btransient activity against C. giveni, cno activity against C. giveni, transient activity against Pyronota species, dtransient activity against Pyronota spp., e Pyronota-active. Figure modified from Hurst et al. (Citation2021).
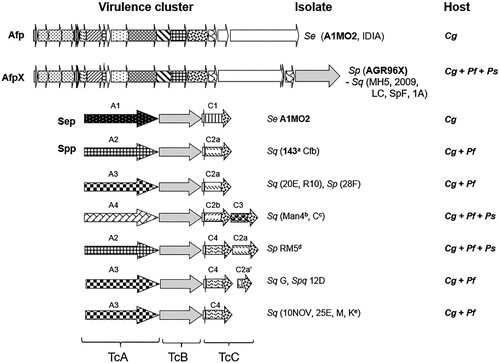
A novel S. proteamaculans isolate AGR96X was found to consistently kill larvae of both C. giveni and Pyronota spp. faster than other isolates, across multiple bioassays over different seasons. AGR96X challenged C. giveni and Pyronota spp. larvae died within 5–12 days of ingestion. Unlike S. entomophila, AGR96X was found to invade the C. giveni larval haemocoelic cavity within days where it rapidly multiplied to reach 8.2 × 109 bacteria per larva, compared to 2.7 × 107 of S. entomophila while confined to the gut at 6 days post challenge (Hurst et al. Citation2018). The main virulence determinant of AGR96X is a variant of the Afp designated AfpX () which, unlike the Afp, contains two tail-length termination proteins (AfpX16a and AfpX16b), two predicted toxins (AfpX17 and AfpX18) and differs in its Afp13 receptor binding tail fibres. Like AGR96X, the purified AfpX is also active towards C. giveni and Pyronota spp. larvae, causing a similar disease phenology (Hurst et al. Citation2018). The broader host range, with consistent increased speed of kill, suggests that AGR96X could be useful in biopesticide formulations in pasture systems.
Ecology and epidemiology of the host specific pathogen Serratia entomophila
Serratia spp. are a common component of the soil microflora which can contain up to 109 bacteria/g soil. The study of the ecology of Serratia species is aided by their fortuitous ability to grow on otherwise toxic caprylate thallous agar (Starr et al. Citation1976). This enables Serratia spp. to be selectively isolated and quantified from complex microbial communities including clinical samples, dead insects and soil. Once putative Serratia colonies have been isolated on the semi selective caprylate thallous agar, identity of genera and species of interest can be made by sequential plating on DNAse agar, a positive test to confirm Serratia spp., then itaconate agar, selective for S. entomophila; and finally, adonitol agar which is able to differentiate S. entomophila from S. proteamaculans (O’Callaghan and Jackson Citation1993a). Virulence of isolates is subsequently confirmed by bioassay (Jackson and Saville Citation2000). Using this selection regime, Serratia species populations in the soil can be quantified and compared to populations and life stages of C. giveni.
Serratia populations, particularly S. entomophila, are generally absent from new pastures but are common where C. giveni is established (O’Callaghan et al. Citation1999). Serratia populations in C. giveni colonised pasture soils averaged 5 × 104 bacteria/g soil but the distribution within pastures was uneven and populations as high as 107 bacteria/g soil were recorded in locations with high C. giveni density. High density patches of C. giveni favour transmission between larvae. Natural infections with >50% amber disease have been recorded (Jackson et al. Citation1991) and a single larva can contribute > 3 × 107 S. entomophila cells (Jackson et al. Citation2001; Hurst et al. Citation2018) to the soil on decay of the cadaver. Accordingly, after a cycle of disease the numbers of pathogenic Serratia in soil increase during June-July and amber disease in C. giveni is more prevalent in high density C. giveni populations, where host proximity favours transmission (O’Callaghan et al. Citation1999). To establish new foci of infection, it appears that pathogenic S. entomophila is dispersed from infected sites by adult beetles during the flight season (O’Callaghan and Jackson Citation1993b). This scenario would account for the dispersal of Serratia during the beetle flight season and colonisation of areas where the pathogen is absent (Zydenbos et al. Citation2016).
The field density of amber diseased C. giveni larvae will, to a large extent, determine the number of Serratia available to cause disease in the subsequent season (Godfray et al. Citation1999). Grass grub populations typically increase over a 5-year period before undergoing a natural decline as the incidence of disease increases in the grub population, concurrent with increased S. entomophila numbers in soil (O’Callaghan et al. Citation1999). Following collapse of the host population, pathogen levels decline allowing the host population to recover in a delayed, density-dependent oscillation until the density is such that transmission is favoured, and a further epizootic occurs (Jackson et al. Citation2017). In the absence of a host, amber disease-causing isolates of Serratia decline resulting in the majority of Serratia isolates recovered from soil being non-pathogenic (O'Callaghan et al. Citation1999).
These pathogen–host dynamics may be affected by external influences such as climate. Under laboratory conditions, S. entomophila populations rapidly declined in dry soils with the decline accentuated when the soil temperature increased from 10°C to 20°C (O’Callaghan et al. Citation2001). The impact of desiccation stress on Serratia populations and other grass grub pathogens in soil can also be inferred from field observations where grass grub outbreaks in pasture commonly occur after dry summers (Barlow and Jackson Citation1998).
The seasonal increase in soil populations and the maintenance of Serratia populations across years may also result from saprophytic growth. Non-pathogenic modes of reproduction have been reported in other entomopathogens and are common for many disease-causing bacteria in mammals (e.g. E. coli). Serratia spp. are capable of rapid growth in soil in the absence of competition with soil microorganisms (for example in sterile soil) and in response to addition of nutrients to soil (O’Callaghan et al. Citation1988). Godfray et al. (Citation1999) constructed a model that incorporated saprophytic growth of the bacteria and illustrated the benefit of a saprophytic phase in Serratia – C. giveni population dynamics.
Use of S. entomophila for management of Costelytra giveni
The natural impact of amber disease on populations of C. giveni suggested that the causative bacterium, S. entomophila, could be used as an applied agent for control of the pest (Jackson Citation1984). The bacterium was cultured by fermentation and delivered in an aqueous drench to field populations of C. giveni resulting in 30–59% infection within three months and residual activity (Jackson et al. Citation1986). Cessation of larval feeding allowed a 30% increase in pasture production over the test period. However, the large volume of liquid inoculant and the sensitivity of bacteria to UV light on the soil surface limited the practicality of the approach. These problems were overcome with development of a high bacterial density aqueous formulation which was injected into the soil through a modified seed drill (O’Callaghan et al. Citation1987). After further field efficacy and safety testing the bacterial product was registered and marketed as ‘Invade’ (Jackson et al. Citation1992). The limitations of this approach (the need for specialist application equipment and refrigerated storage) were overcome with development of an ambient temperature stable granular formulation of the bacteria which could be applied through standard drills which was marketed as ‘Bioshield’ (Johnson et al. Citation2001; Jackson Citation2007). When compared with a chemical insecticide (diazinon) for long term management of C. giveni (Zydenbos et al. Citation2016) application of S. entomophila established a cycle of disease in the larval population and maintained the population at low levels for several seasons. In contrast, the insecticide treatment caused a rapid decline in larval numbers shortly after application, but after two years the insect population resurged to high, damaging levels.
Successful use of S. entomophila in control of C. giveni requires consideration of the characteristics of the pest and the bacterium. Healthy populations of the beetles multiply rapidly in new pastures resulting in a damage peak frequently observed 2–3 years post pasture establishment. S. entomophila requires a basal population of insects sufficient for multiplication and transmission. If applied to a rising population a level of 20% infection within six weeks of application is usually sufficient to establish a recycling epizootic of amber disease-causing bacteria which will hold the pest population to low levels (Jackson et al. Citation2017). As a biological control agent, S. entomophila is limited by its specificity to a single pest (Jackson et al. Citation1991) and its need for accurate application timing; these are characteristics borne out of close evolution with its host.
Epidemiology of non-amber disease Serratia pathotypes
Although the epidemiology of S. entomophila and C. giveni are reasonably well understood, the same is not true of atypical disease causing Serratia pathotypes. It is likely that faster acting pathogens such as S. proteamaculans AGR96X may be associated with localised disease outbreaks characterised by a ‘boom to bust’ life cycle or, alternately, suited to cooler climes where reduced larval metabolism and movement limits spread of the pathogen (Read Citation1994; Paez and Fleming-Davies Citation2020). Unlike S. entomophila, AGR96X invades the haemoceol cavity early in the infection and can reach cell numbers of greater than 8 × 109 cells per larva. This, combined with mortality and decay within 12 days of infection, will increase the opportunity for recycling of AGR96X through infection of heathy larvae within the season. The ability of some S. proteamaculans or S. quinivorans isolates to cause disease in 60–90% of the challenged larvae (Hurst et al. Citation2021) may reflect differences in virulence regulation and/or larval genetics and physiology. In this scenario the unaffected larvae may ensure the long-term persistence of the pathogen by maintaining availability of the host in subsequent years.
The diversity of S. proteamaculans and S. quinivorans isolates with activity against both C. giveni and P. festiva likely reflects the overlapping distribution of these insect species. Amber disease-encoding isolates along with other Serratia Tc-encoding variants have been mainly isolated from low-lying regions of modified pasture (typically comprising clover and ryegrass) inhabited by C. giveni and/or Pyronota species (Jackson et al. Citation1991; Dodd et al. Citation2006; Hurst et al. Citation2021), although only limited sampling has been conducted to date. The most virulent Serratia isolates (e.g. strains encoding AfpX) were recovered from modified pastures in hill country areas of Aotearoa. In addition, both the AfpX encoding AGR96X and the broad host-range entomopathogen Y. entomophaga (Hurst, Becher, Young, et al. Citation2011a) were isolated from the same field site (Hurst et al. Citation2021), indicative of a multi-entomopathogen complex at this location. This site was at elevation and in proximity to the native estate wherein potential pathogen spill-over from areas of endemic flora comprising tussock and other plant species may have occurred. Although yet to be defined, aside from the likely temperature differences (low land versus sub alpine -possibly affecting larval metabolism), the different plant species present in the native estate or modified pasture (clover and ryegrass) may impart positive/negative selective pressures for S. entomophila and the various species of S. proteamaculans or S. quinivorans. It is plausible that, as previously noted with the endophytic nature of S. proteamaculans isolate 568 (Taghavi et al. Citation2009), some Aotearoa Serratia isolates may be plant associated.
Origins of pathogenicity of Serratia spp. to scarabs within Aotearoa
In pre-human Aotearoa, C. giveni and Pyronota spp. probably lived in tussock patches on the edge of the native bush or in areas temporarily cleared and revegetated after avalanche or flood (much as is seen in the high country today). In these situations, a balance develops between pathogen and host. With human intervention through agriculture, soil disturbance during cultivation can virtually eliminate pests and with them their natural pathogens, explaining the healthy state of scarab beetles colonising new pastures. In the depauperate new pasture environment C. giveni populations increase without constraint. The absence of pathogens leads to spectacular pest outbreaks in pasture, especially after dramatic land use change (Jackson et al. Citation2012). C. giveni and Pyronota spp. are usually univoltine and females can have an egg load of up to 60. For both C. giveni and P. setosa most eggs are laid close to where the female emerges leading to high density, pasture damaging, patches of larvae.
The bacterial pathogens of Aotearoa scarabs have evolved from the Gammaproteabacteria widespread on Earth nearly 2 billion years ago. Bacteria evolved together with other life forms and formed associations with invertebrates, some of which were pathogenic. Evidence of ancient relationships is provided by P. popilliae and P. lentimorbus which are only known to infect members of the coleopteran family Scarabaeidae, including those in Aotearoa. The precursors of the current scarab beetle species are known from the fossil record in the Mesozoic period, before the breakup of the Pangaea supercontinent about 175 million years ago (Morón Citation1984; Ahrens et al. Citation2014). Scarab beetles can be found on all habitable continents and, with them, infections of Paenibacillus spp. which appear to show host specificity (Klein and Jackson Citation1992), suggesting that they co-evolved with the beetles and emerged from ancient origins before continental break-up. The suggestion of ancient origins is supported by molecular evidence from Zeigler (Citation2013). Similarly, the unique associations of Photorhabdus spp. bacteria with Heterorhabditis spp. nematodes and Xenorhabdus spp. bacteria with Steinernema spp. nematodes from all continents suggests coevolution before the Pangaea separation (Poinar Citation2011; Poinar and Grewal Citation2012). The nematode/Photorhabdus association is of interest as this suggests that Tc genes, or their precursors, were widespread before the breakup of Pangea with origins in the Mesozoic and have evolved into the insect pathogens of the Xenorhabdus (Sergeant et al. Citation2006), Photorhabdus (Duchaud et al. Citation2003), Yersinia (Fuchs et al. Citation2008) and Serratia genera of the Enterobacteriaceae (Jackson et al. Citation2017).
The pADAP encoded sep and afp genes together with the high specificity of isolates causing amber disease of C. giveni suggest that this is a more recently evolved host–pathogen relationship. Bacterial evolution is aided by fluid genomes adapting by mutation and homologous recombination with the potential for plasmid transfer of genetic material between cells. For the toxin-bearing B. thuringiensis, strain evolution has probably been driven quite rapidly with exchange of toxin-coding plasmids (Bizzarri and Bishop Citation2008) such that there are a huge range of strains of B. thuringiensis in the environment containing functionally different toxins (Cry, Cyt, VIP) governing pathogenicity to different insect groups (Jurat-Fuentes and Jackson Citation2012). The diversity of Tc-encoding clusters across Proteobacteria signifies that the Tc proteins play a key role in the ecology of these bacteria (Song et al. Citation2021) and alludes to their potential mobility. It may also reflect an enhanced evolutionary capacity for these Tc encoding regions to acquire or interchange different tcC components, as originally proposed by Hill et al. (Citation1994). The diversity of Tc encoding clusters parallels the diversity of three-domain Cry toxins in B. thuringiensis, where the various composite domains are proposed to have evolved at their own rates (Bravo et al. Citation2013), with greater variability in the host-targeting and effector regions (Wu et al. Citation2007). In this instance the varying TcA, Afp13 cell targeting, and TcC effector regions (Hurst et al. Citation2000; Hurst et al. Citation2004; Hurst et al. Citation2021) have likely evolved to target and/or have greater affinity for certain host species. This selective process, in turn, may be driven by a pathogen–host arms race as has been proposed in other effector systems (Galan Citation2009; Białas et al. Citation2018). Given the variability within Tc orthologs and associated TcC effectors, it remains unclear as to why there are only two main Afp variants (Afp and AfpX).
Sitter et al. (Citation2021) and Williams et al. (Citation2022) considered that pADAP-type plasmids are restricted to genus Serratia. To date the pathogenicity encoding genes for the Aotearoa Scarabaeidae have been defined as plasmid located (Sitter et al. Citation2021). It is therefore tempting to speculate that the various STAMPs and their associated toxins likely reflect the ecology of the pathogen and the host. None of the non amber disease encoding strains of S. proteamaculans or S. quinivorans encoded both a tc and an afp variant. This may reflect that rapid onset of disease imparted by these isolates overrides the requirement for a second virulence determinant. Alternately, the cessation of feeding activity imparted by the Afp may necessitate the requirement for Sep proteins to clear the gut. In turn the voided gut may make intestinal conditions more favourable to pathogen establishment.
The host specificity of the various S. proteamaculans, S. quinivorans and S. entomophila strains, combined with the absence of non-Aotearoa STAMP variants in the current public bioinformatic databases, supports the notion that the assembly of STAMPs has been essential for success of the disease since the separation of Aotearoa from Gondwanaland. Bioinformatic analysis of the various STAMP variants revealed that the key virulence determinants have been independently acquired in a distinct region of the STAMP backbone (Hurst et al. Citation2011b; Sitter et al. Citation2021). The association of STAMPs within Serratia spp., wherein S. entomophila is more geographically localised (Aotearoa), is indicative of more recent evolution. The isolation of an avirulent S. proteamaculans pADAP variant pPuna18 from Aotearoa, encoding a nitrogen fixation cluster, further suggests that the pADAP linage is diverse within Aotearoa and may be confined to S. entomophila, S. quinivorans, S. proteamaculans, and S. liquefaciens (Sitter et al. Citation2021). It is likely that the precursors of the Sep and Afp toxins were present among the Enterobacteriacae and acquired in Serratia spp. to form a primitive STAMP. Further to this the altered virulence of S. proteamaculans isolate 3041 transconjugant containing the plasmid p145, was less virulent than the S. proteamaculans 145, plasmid p145 donor. This finding suggests that the chromosomal background affects disease regulation and that these plasmids are likely confined to an isolate lineage (Sitter et al. Citation2021).
Relative to other genera, the pan genome of Serratia is considered restrained and metabolically diverse, indicative of long-term niche adaptation within evolutionary time scales (Williams et al. Citation2022). The S. entomophila chromosome has been defined as ‘closed’, with reduced signatures of horizontal gene transfer, pointing to speciation at the time that pADAP was acquired (Vaughan et al. Citation2022; Williams et al. Citation2022). Of interest, S. entomophila encodes the species-specific itaconate encoding region. An itaconate-deleted mutant resulted in a delay in the onset of amber disease indicating a role of this metabolic pathway in the initial disease process and perhaps a tenuous link to speciation (Vaughan et al. Citation2022).
The pathogenesis of amber disease caused by S. entomophila with these toxins (Sep and Afp) in Aotearoa scarabs is unusual (Jackson et al. Citation1993; Jackson et al. Citation2001) in that there is no observable tissue damage in the early stages of infection but there are physiological responses (cessation of feeding and gut clearance) caused by combination of expression of the Sep and Afp toxins. As previously noted, a continuous supply of Sep proteins is required to maintain gut clearance, while an oversupply of Afp causes larval mortality. This, combined with elevated chitinase levels in a S. entomophila QS mutant (Hurst Citation2016), suggests that gene regulation within this bacterium is tightly regulated to prolong the amber disease process. In this scenario, under conditions of high cell density such as can occur in the C. giveni hindgut, chitinase production is likely decreased, reducing the ability of S. entomophila to degrade the peritrophic membrane or other chitinaceous material. This, in turn, may increase extend the time required before the bacteria can invade the haemocoelic cavity, thereby potentially extending the persistence of the pathogen by protecting it from environmental stresses in the soil (Hurst Citation2016).
Recombinations and/or genetic acquisitions of the S. proteamaculans/S. quinivorans plasmid by S. entomophila probably led to the variant with the capability and evolutionary advantages of infection and growth in scarab larvae. Chance association and plasmid transfer would have conferred this advantage on S. entomophila, which is a relatively rare bacterium in the non-pathogenic form, both internationally and in Aotearoa pasture soils. With European colonisation and wide scale pasture development in Aotearoa, the pasture scarabs and their Serratia pathogens have proliferated to become common features of the local environment. This is most evident with the host specific S. entomophila – C. giveni relationship and the chronic nature of amber disease where the prolonged infection is suggestive of a dedicated pathogen with a lifestyle somewhat dependent on its C. giveni host.
Conclusion
The isolation of Serratia strains encoding different Tc and Afp clusters offers the potential for development of biopesticides that are more efficacious in specific environments. Further, combining isolates with different virulence properties, such as S. entomophila and the more rapid-killing S. proteamaculans isolates may afford greater control of the insect host (Paez and Fleming-Davies Citation2020). This, combined with the identification of Serratia pathogens that target C. giveni larvae with differing effects, suggests genetic heterogeneity within C. giveni across different geographies. This may in part explain the presence of several different Tc encoding clusters. To validate these scenarios detailed genetic analysis of the C. giveni larvae sourced from different locations needs to be undertaken. Such studies will be aided through advances in metagenomic and target specific quantitative assays which will enable the differentiation of pathogen and host type across both the natural and modified Aotearoa landscapes.
Acknowledgements
We thank Leo Eberl University of Zurich for assessment of AHL production of the S. entomophila QS mutant.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Ahrens D, Schwarzer J, Vogler AP. 2014. The evolution of scarab beetles tracks the sequential rise of angiosperms and mammals. Proc R Soc. B. 2812014147020141470. doi:10.1098/rspb.2014.1470.
- Ashelford KE, Fry JC, Bailey MJ, Day MJ. 1983. Characterization of Serratia isolates from soil, ecological implications and transfer of Serratia proteamaculans subsp. quinovora Grimont et al. 1983 to Serratia quinivorans corrig., sp. nov. Int J Syst Evol Microbiol. 52(6):2281–2289. doi:10.1099/00207713-52-6-2281.
- Barlow ND, Jackson TA.. 1998. A probabilistic model for augmentative biological control of grass grub (Costelytra zealandica) in Canterbury. Proceedings from the 6th Australasian Grassland Invertebrate Ecological Conference 199. p. 290- 296.
- Begrem S, Jérôme M, Leroi F, Delbarre-Ladrat C, Grovel O, Passerini D. 2021. Genomic diversity of Serratia proteamaculans and Serratia liquefaciens predominant in seafood products and spoilage potential analyses. Int J Food Microbiol. 354:109326. doi:10.1016/j.ijfoodmicro.2021.109326.
- Białas A, Zess EK, De la Concepcion JC, Franceschetti M, Pennington HG, Yoshida K, Upson JL, Chanclud E, Wu CH, Langner T, et al. 2018. Lessons in effector and NLR biology of plant-microbe systems. Mol Plant Micro Interact. 31(1):34–45. doi:10.1094/MPMI-08-17-0196-FI.
- Bizzarri MF, Bishop AH. 2008. The ecology of Bacillus thuringiensis on the phylloplane: colonisation from soil, plasmid transfer, and interaction with larvae of Pieris brassicae. Microb Ecol. 56(1):133–139. doi:10.1007/s00248-007-9331-1.
- Blackburn MB, Martin PAW, Kuhar D, Farrar RR Jr, Gundersen-Rindal DE. 2011. The occurrence of Photorhabdus-like toxin complexes in Bacillus thuringiensis. PLoS ONE. 6:e18122. doi:10.1371/journal.pone.0018122.
- Böck D, Medeiros JM, Tsao HF, Penz T, Weiss GL, Aistleitner K, Horn M, Pilhofer M. 2017. In situ architecture, function, and evolution of a contractile injection system. Science. 357(6352):713–717. doi:10.1126/science.aan7904.
- Bourner TC, Glare TR, O’Callaghan M, Jackson TA. 1996. Towards greener pastures – pathogens and pastures. NZ J Ecol. 20(1):101–107. https://newzealandecology.org/nzje/1992.
- Bowen D, Rocheleau TA, Blackburn M, Andreev O, Golubeva E, Bhartia R, ffrench-Constant RH. 1998. Insecticidal toxins from the bacterium Photorhabdus luminescens. Science. 280(5372):2129–2132. doi:10.1126/science.280.5372.2129.
- Bravo A, Gomez I, Porta H, Garcia-Gomez BI, Rodriguez-Almazan C, Pardo L, Soberon M. 2013. Evolution of Bacillus thuringiensis Cry toxins insecticidal activity. Microb Biotech. 6(1):17–26. doi:10.1111/j.1751-7915.2012.00342.x.
- Brown JG. 1963. Biology and taxonomy of the genus Pyronota Bois. (Melolonthinae, Coleoptera). University of Canterbury (Lincoln College).
- Brunner-Mendoza C, Reyes-Montes MR, Moonjely S, Bidochka MJ, Toriello C. 2019. A review on the genus Metarhizium as an entomopathogenic microbial biocontrol agent with emphasis on its use and utility in Mexico. Biocontrol Sci Tech. 29(1):83–102. doi:10.1080/09583157.2018.1531111.
- Busby JN, Panjiker S, Landsberg MJ, Hurst MRH, Lott SJ. 2013. The BC component of ABC toxins is an RHS-repeat-containing protein encapsulation device. Nature. 501:547–550. doi:10.1038/nature12465.
- Chattopadhyay P, Chatterjee S, Gorthi S, Sen SK. 2012. Exploring agricultural potentiality of Serratia entomophila AB2: dual property of biopesticide and biofertilizer. British Biotechnol J. 2:1–12. doi:10.9734/BBJ/2012/778.
- Chattopadhyay P, Gorthi S, Chatterjee S, Sen SK. 2011. Characterization of bacterial isolates as natural biocontrolling agents of bollworm from an epizootic pest (Heliothis armigera). Pest Tech. 5:81–85.
- Chen L, Song N, Liu B, Zhang N, Alikhan N-F, Zhou Z, Zhou Y, Zhou S, Zheng D, Chen M, et al. 2019. Genome-wide identification and characterization of a superfamily of bacterial extracellular contractile injection systems. Cell Reports. 29(2):511–521.e2. doi:10.1016/j.celrep.2019.08.096.
- Coca-Abia MM, Romero-Samper J. 2016. Establishment of the identity of Costelytra zealandica (White 1846) (Coleoptera: Scarabaeidae: Melolonthinae) a species commonly known as the New Zealand grass grub. NZ Entomol. 39(2):129–146. doi:10.1080/00779962.2016.1230254.
- Desfosses A, Venugopal H, Joshi T, Felix J, Jessop M, Jeong H, Hyun J, Heymann JB, Hurst MRH, Gutsche I, Mitra AK. 2019. Atomic structures of an entire contractile injection system in both the extended and contracted states. Nat Microbiol. 4:1885–1894. doi:10.1038/s41564-019-0530-6.
- Dodd SJ, Hurst MRH, Glare TR, O'Callaghan M, Ronson CW. 2006. Occurrence of Sep insecticidal toxin complex genes in Serratia species and Yersinia frederiksenii. Appli Environ Microbiol. 72(10):6584–6592. doi:10.1128/AEM.00954-06.
- Duchaud E, Rusniok C, Frangeul L, Buchrieser C, Givaudan A, Taourit S, Bocs S, Boursaux-Eude C, Chandler M, Charles JF, et al. 2003. The genome sequence of the entomopathogenic bacterium Photorhabdus luminescens. Nat Biotechnol. 21:1307–1313. doi:10.1038/nbt886.
- Dunbar JE, Hateley PW, Townsend RJ, Zydenbos SM, Jackson TA. 2012. Assessing damage by Manuka beetles (Pyronota spp) in flipped west coast pastures. NZ Plant Pro Soc. 65:295–295. doi:10.30843/nzpp.2012.65.5410.
- Ferguson CM, Barratt BIP, Bell N, Goldson SL, Hardwick S, Jackson M, Jackson TA, Phillips CB, Popay AJ, Rennie G, et al. 2019. Quantifying the economic cost of invertebrate pests to New Zealand’s pastoral industry. NZ J Agri Res. 62(3):255–315. doi:10.1080/00288233.2018.1478860.
- ffrench-Constant R, Waterfield N. 2005. An ABC guide to the bacterial toxin complexes. Adv Appl Microbiol. 58C:169–183. doi:10.1016/S0065-2164(05)58005-5.
- Fuchs TM, Bresolin G, Marcinowski L, Schachtner J, Scherer S. 2008. Insecticidal genes of Yersinia spp.: taxonomical distribution, contribution to toxicity towards Manduca sexta and Galleria mellonella, and evolution. BMC Microbiol. 8:214. doi:10.1186/1471-2180-8-214.
- Galan JE. 2009. Common themes in the design and function of bacterial effectors. Cell Host Microbe. 5(6):571–579. doi:10.1016/j.chom.2009.04.008.
- Gatehouse HS, Marshall SDG, Simpson RM, Gatehouse LN, Jackson TA, Christeller JT. 2008. Serratia entomophila innoculation causes a defect in exocytosis in Costelytra zealandica larvae. Insect Mol Biol. 17(4):375–385. doi:10.1111/j.1365-2583.2008.00807.x.
- Gatehouse HS, Tan B, Christeller JT, Hurst MRH, Marshall SDG, Jackson TA. 2009. The relationship between amber disease phenology and digestive enzymes in larvae of the New Zealand grass grub (Costelytra zealandica). J Invert Path. 101(3):215–221. doi:10.1016/j.jip.2009.05.010.
- Gatsogiannis C, Lang AE, Meusch D, Pfaumann V, Hofnagel O, Benz R, Aktories K, Raunser S. 2013. A syringe-like injection mechanism in Photorhabdus luminescens toxins. Nature. 495:520–523. doi:10.1038/nature11987.
- Given BB, Hoy JM. 1952. A revision of the Melolonthinae of New Zealand. NZ DSIR Ind Res Bull No 102.
- Glare TR, Corbett GE, Sadler AJ. 1993. Association of a large plasmid with amber disease of the New Zealand grass grub, Costelytra zealandica, caused by Serratia entomophila and Serratia proteamaculans. J Invertebr Pathol. 62(2):165–170. doi:10.1006/jipa.1993.1091.
- Glare TR, O’Callaghan M, Wigley PJ. 1993. Checklist of naturally occurring entomopathogenic microbes and nematodes in New Zealand. NZ J Zool. 20(2):95–120. doi:10.1080/03014223.1993.10422867.
- Glare TR, Scholte O, Reimer YSO, Cummings N, Rivas-Franco F, Nelson TL, Zimmermann G. 2021. Diversity of the insect pathogenic fungi in the genus Metarhizium in New Zealand. NZ J Botany. 59(4):440–456. doi:10.1080/0028825X.2021.1890155.
- Godfray HCJ, Briggs CJ, Barlow ND, O’Callaghan M, Glare TR, Jackson TA. 1999. A model of insect—pathogen dynamics in which a pathogenic bacterium can also reproduce saprophytically. Proc. of the Royal Society London B. 266:233–240. doi:10.1098/rspb.1999.0627.
- Grebennikov Vasily V, Scholtz Clarke H. 2004. The basal phylogeny of Scarabaeoidea (Insecta : Coleoptera) inferred from larval morphology. Invert System. 18(3):321–348. doi:10.1071/IS03013.
- Grimont F, Grimont PAD. 2006. The genus Serratia. Prokaryotes. 6:219–244. doi:10.1007/0-387-30746-X_11.
- Grimont PAD, Grimont F, Starr MP. 1981. Serratia species isolated from plants. Curr Microbiol. 5:317–322. doi:10.1007/BF01567926.
- Grimont PAD, Jackson TA, Ageroe E, Noonan MJ. 1988. Serratia entomophila sp. nov. associated with amber disease in the New Zealand grass grub Costelytra zealandica. Int J Syst Bacteriol. 38(1):1–6. doi:10.1099/00207713-38-1-1.
- Grkovic S, Glare TR, Jackson TA, Corbett GE. 1995. Genes essential for amber disease in grass grubs are located on the large plasmid found in Serratia entomophila and Serratia proteamaculans. Appl Environ Microbiol. 61(6):2218–2223. doi:10.1128/aem.61.6.2218-2223.1995.
- Hill CW, Sandt CH, Vlazny DA. 1994. Rhs elements of Escherichia coli: a family of genetic composites each encoding a large mosaic protein. Mol Microbiol. 12(6):865–871. doi:10.1111/j.1365-2958.1994.tb01074.x.
- Hurst MR, Glare TR, Jackson TA. 2004. Cloning Serratia entomophila antifeeding genes a putative defective prophage active against the grass grub Costelytra zealandica. J Bacteriol. 186(15):5116–5128. doi:10.1128/JB.186.15.5116-5128.2004.
- Hurst MR, Glare TR, Jackson TA, Ronson CW. 2000. Plasmid-located pathogenicity determinants of Serratia entomophila, the causal agent of amber disease of grass grub, show similarity to the insecticidal toxins of Photorhabdus luminescens. J Bacteriol. 182(18):5127–5138. doi:10.1128/JB.182.18.5127-5138.2000.
- Hurst MRH. 2016. Non-spore-forming bacterial entomopathogens: Their toxins, hosts and the environment: why be a pathogen. In: Hurst C, editor. The Rasputin effect: when commensals and symbionts become parasitic. Vol. 3. Advanc Environ Microbiol. Cham: Springer; p. 169–220.
- Hurst MRH, Beard SS, Jackson TA, Jones SM. 2007. Isolation and characterization of the Serratia entomophila antifeeding prophage. FEMS Microbiol Lett. 270(1):42–48. doi:10.1111/j.1574-6968.2007.00645.x.
- Hurst MRH, Beattie A, Jones SA, Laugraud A, van Koten C, Harper L. 2018. Serratia proteamaculans strain AGR96X encodes an antifeeding prophage (Tailocin) with activity against grass grub (Costelytra giveni) and manuka beetle (Pyronota species) larvae. Appl Environ Microbiol. 84:e02739-17. doi:10.1128/AEM.02739-17.
- Hurst MRH, Beattie A, Laugraud A, Townsend R, Sitter L, van Koten C, Harper L. 2021. Identification of diverse toxin complex clusters and an eCIS variant in Serratia proteamaculans pathovars of the New Zealand grass grub (Costelytra giveni) and Manuka beetle (Pyronota spp.) larvae. Microbiol Spectrum. doi:10.1128/Spectrum.01123-21.
- Hurst MRH, Becher A, O’Callaghan M. 2011b. Nucleotide sequence of the Serratia entomophila plasmid pADAP and the Serratia proteamaculans pU143 plasmid virulence associated region. Plasmid. 65(1):32–41. doi:10.1016/j.plasmid.2010.10.001.
- Hurst MRH, Becher SA, Young SD, Nelson TL, Glare TR. 2011a. Yersinia entomophaga sp. nov. isolated from the New Zealand grass grub Costelytra giveni. Int J Syst Bact Evol Micro. 61(4):844–849. doi:10.1099/ijs.0.024406-0.
- Hurst MRH, Jackson TA. 2002. Use of green fluorescent protein to monitor the fate of Serratia entomophila causing amber disease in the New Zealand grass grub, Costelytra zealandica. J Microbiol Methods. 50(1):1–8.doi: 10.1016/s0167-7012(02)00004-0.
- Hurst MRH, Jones S, Tan B, Jackson TA. 2007. Induced expression of the Serratia entomophila Sep proteins shows activity towards the larvae of the New Zealand grass grub Costelytra zealandica. FEMS Microbiol Lett. 275(1):160–167. doi:10.1111/j.1574-6968.2007.00886.x.
- Hurst MRH, Richards NK, Brown R, Mansfield S. 2022. Grass grub is spreading on the South Island’s West Coast. NZ Plant Pro Soc. 75:9–11.
- Jackson T, Berry C, O'Callaghan M. 2017. Chapter 8. Bacteria. In: Hajek AE, Shapiro-Ilan DI, editors. Ecology of invertebrate diseases. Hoboken: Wiley; p. 287–326.
- Jackson TA. 1984. Honey disease, an indicator of population decline in grass grub. Proceedings of the 37th NZ Weed and Pest Conf. 33:113–116. doi:10.30843/nzpp.1984.37.9519.
- Jackson TA. 1995. Amber disease reduces trypsin activity in midgut of Costelytra zealandica Coleoptera; Scarabaeidae larvae. J Invert Path. 65(1):68–69. doi:10.1006/jipa.1995.1009.
- Jackson TA. 2003. Environmental safety of inundative application of a naturally occurring biocontrol agent, Serratia entomophila. In: Hokkanen HMT, Hajek AE, editors. Environmental impacts of microbial insecticides. Progress in biological control. Vol. 1. Dordrecht: Springer. doi:10.1007/978-94-017-1441-9_9.
- Jackson TA. 2007. A novel bacterium for control of grass grub. In: Vincent C, Goettel MS, Lazarovits G, editors. Biological control: a global perspective. Wallingford: CABI publishing; p. 160–168.
- Jackson TA, Boucias DG, Thaler JO. 2001. Pathobiology of amber disease, caused by Serratia spp., in the New Zealand grass grub, Costelytra zealandica. J Invertebr Path. 78(4):232–243. doi:10.1006/jipa.2002.5078.
- Jackson TA, Glare TR, O'Callaghan M. 1991. Pathotypic boundaries for Serratia spp. causing amber disease in the New Zealand grass grub, Costelytra zealandica. In: Smits PH, editor. Proceedings of the 3rd European Meeting of Microbial Control of Pests. International Organization for Biological Control of Noxious Plants and Animals, Wageningen, The Netherlands, p. 148–152.
- Jackson TA, Huger AM, Glare TR. 1993. Pathology of amber disease in the New Zealand grass grub, Costelytra zealandica Coleoptera: Scarabaeidae. J Invert Path. 61(2):123–130. doi:10.1006/jipa.1993.1024.
- Jackson TA, O’Callaghan M, Glare TA. 1998. Safe use of replicating bacteria in biological control. Phytoprotection 79 (Suppl.) OECD Workshop, sustainable pest management, safe utilization of new organisms in biological control; Sept 27–30, 1997; Montreal Quebec, Canada. p. 50–55. doi:10.7202/706157ar.
- Jackson TA, Pearson JF, O'Callaghan M, Mahanty HK, Willocks M. 1992. Pathogen to product – development of Serratia entomophila Enterobacteriaceae as a commercial biological control agent for the New Zealand grass grub Costelytra zealandica. In: Jackson TA, Glare T, editors. Use of pathogens in scarab pest management. Andover: Intercept Ltd. p. 191–198.
- Jackson TA, Pearson JF, Stucki G. 1986. Control of the grass grub, Costelytra zealandica (white) (Coleoptera: Scarabaeidae), by application of the bacteria Serratia spp. causing honey disease. Bull Entomol Res. 76:69–76. doi:10.1017/S0007485300015297.
- Jackson TA, Saville DJ. 2000. Bioassays of replicating bacteria against soil-dwelling insect pests. In: Asher KRS, Navon A, editors. Bioassays of entomopathogenic microbes and nematodes. New York (N.Y.): CAB International. p. 73–94.
- Jackson TA, Townsend RJ, Dunbar JE, Ferguson CM, Marshall SDG, Zydenbos SM. 2012. Anticipating the unexpected- managing pasture pest outbreaks after large-scale land conversion. Proc NZ Grass Assoc. 74:147–152. doi:10.33584/jnzg.2012.74.2861.
- Johnson VW, Pearson JF, Jackson TA. 2001. Formulation of Serratia entomophila for biological control of grass grub. NZ Plant Protect. 54:125–127. doi:10.30843/nzpp.2001.54.3752.
- Jurat-Fuentes JL, Jackson TA. 2012. Bacterial pathogens. In: Vega FE, Kaya HK, editors. Insect pathology. 2nd ed. London: Academic Press, Elsevier; p. 265–349.
- Klein MG, Grewal PS, Jackson TA, Koppenhöfer AM. 2007. Lawn, turf and grassland pests. In: Lacey LA, Kaya HK., editors. Field manual of techniques in invertebrate pathology. Dordrecht: Springer; p. 681–706.
- Klein MG, Jackson TA. 1992. Bacterial diseases of scarabs. In: Jackson TA, Glare TR, editors. Use of pathogens in scarab pest management. Andover: Intercept; p. 43–61.
- Lefort MC, Worner SP, De Romans S, Armstrong K, Glare TR, Boyer S. 2014. Invasion success of a scarab beetle within its native range: host range expansion vs. host-shift. PeerJ. 2:e262. doi:10.7717/peerj.262.
- Meusch D, Gatsogiannis C, Efremov RG, Lang AE, Hofnagel O, Vetter IR, Aktories K, Raunse S. 2014. Mechanism of Tc toxin action revealed in molecular detail. Nature. 508:61–65. doi:10.1038/nature13015.
- Morgan JA, Sergeant M, Ellis D, Ousley M, Jarrett P. 2001. Sequence analysis of insecticidal genes from Xenorhabdus nematophilus PMFI296. Appl Environ Microbiol. 67(5):2062–2069. doi:10.1128/AEM.67.5.2062-2069.2001.
- Morón MA. 1984. Escarabajos 200 Milliones de Años de Evolución. Instituto de Ecología. México: pp 132.
- Nunez-Valdez ME, Calderon MA, Aranda E, Hernández L, Ramírez-Gama RM, Lina L, Rodríguez-Segura Z, Gutiérrez MC, Villalobos FJ. 2008. Identification of a putative Mexican strain of Serratia entomophila pathogenic against root damaging larvae of Scarabaeidae (Coleoptera). Appl Environ Microbiol. 74(3):802–810. doi:10.1128/AEM.01074-07.
- O'Callaghan M, Gerard EM, Johnson VW. 2001. Effect of soil moisture and temperature on survival of microbial control agents. NZ Plant Prot Soc. 54:128–135. doi:10.30843/nzpp.2001.54.3753.
- O'Callaghan M, Jackson TA. 1993a. Isolation and enumeration of Serratia entomophila – a bacterial pathogen of the New Zealand grass grub, Costelytra zealandica. J Appl Bacteriol. 75(4):307–314. doi:10.1111/j.1365-2672.1993.tb02781.x.
- O'Callaghan M, Jackson TA. 1993b. Adult grass grub dispersal of Serratia entomophila. Proc 46th NZ Plant Prot Soci Con. Research Note. 46:235–236. doi:10.30843/nzpp.1993.46.11138.
- O'Callaghan M, Noonan MJ, Broad TM, Jackson TA. 1987. Evaluation of methods for applying Serratia entomophila to pasture. Proc 40th NZ Weed and Pest Con Conf. 40:27–30. doi:10.30843/nzpp.1987.40.9938.
- O'Callaghan M, Young SD, Barlow ND, Jackson T. 1999. The ecology of grass grub pathogenic Serratia spp. in New Zealand pastures. In: Mathiessen JN, editor. Proceedings of the 7th Australasian Conference Grassland Invertebrate Ecology, CSIRO Entomol, Perth. p. 85–91.
- O'Callaghan M, Young SD, Barlow ND, Jackson TA. 1988. The ecology of grass grub pathogenic Serratia spp. in New Zealand pastures. In: Proceedings of the 5th Australasian Conference Grassland Invertebrate Ecology. p. 69–75.
- Paez D, Fleming-Davies A. 2020. Understanding the evolutionary ecology of host–pathogen interactions provides insights into the outcomes of insect pest biocontrol. Pest Biocon Viruses. 12(2):141. doi:10.3390/v12020141.
- Poinar GO. 2011. The evolutionary history of nematodes: as revealed in stone, amber and mummies. In: Hunt DJ, Perry RN, editors. Nematology monographs and perspectives. Vol. 9. Leiden-Boston: Brill; p. 429.
- Poinar GO, Grewal PS. 2012. History of entomopathogenic nematology. J Nematol. 44(2):153–161.
- Rangel LI, Henkels MD, Shaffer BT, Walker FL, Davis EW II, Stockwell VO, Bruch D, Taylor BJ, Loper JE. 2016. Characterization of toxin complex gene clusters and insect toxicity of bacteria representing four subgroups of Pseudomonas fluorescens. PLoS ONE. 11(8):e0161120. doi:10.1371/journal.pone.0161120.
- Read A. 1994. The evolution of virulence. Trends Microbiol. 2(3):73–76. doi:10.1016/0966-842X(94)90537-1.
- Rey P, Muller R. 2010. Fragmentation of active continental plate margins owing to the buoyancy of the mantle wedge. Nat Geosci. 3:257–261. doi:10.1038/ngeo825.
- Richards NK, Glare TR, Hall DCA. 1997. Genetic variation in grass grub, Costelytra zealandica, from several regions. NZ Plant Pro Soc. 50:338. doi:10.30843/nzpp.1997.50.11327.
- Rybakova D, Radjainia M, Turner A, Sen A, Mitra AK, Hurst MRH. 2013. Role of antifeeding prophage (Afp) protein Afp16 in terminating the length of the Afp tailocin and stabilizing its sheath. Mol Micro. 89(4):702–714. doi:10.1111/mmi.12305.
- Sarris PF, Ladoukakis ED, Panopoulos NJ, Scoulica EV. 2014. A phage tail-derived element with wide distribution among both prokaryotic domains: a comparative genomic and phylogenetic study. Genome Biol Evol. 6(7):1739–1747. doi:10.1093/gbe/evu136.
- Sergeant M, Baxter L, Jarrett P, Shaw E, Ousley M, Winstanley C, Morgan JAW. 2006. Identification, typing, and insecticidal activity of Xenorhabdus isolates from entomopathogenic nematodes in United Kingdom soil and characterization of the xpt toxin loci. Appl Environ Microbiol. 72(9):5895–5907. doi:10.1128/AEM.00217-06.
- Sitter TL, Vaughan AL, Schoof M, Jackson SA, Glare TR, Cox MP, Finneran PC, Gardner PP, Hurst MRH. 2021. Evolution of virulence in a novel family of transmissible mega-plasmids. Environ Microbiol. 23(9):5289–5304. doi:10.1111/1462-2920.15595.
- Song N, Chen L, Zhou Z, Ren X, Liu B, Zhou S, Wang C, Wu Y, Waterfield NR, Yang J, Yang G. 2021. Genome-wide dissection reveals diverse pathogenic roles of bacterial Tc toxins. PLoS Path. 17(2):e1009102. doi:10.1371/journal.ppat.1009102.
- Starr MP, Grimont PA, Grimont F, Starr PB. 1976. Caprylate-thallous agar medium for selectively isolating Serratia and its utility in the clinical laboratory. J Clin Microbiol. 4(3):270–276. doi:10.1128/jcm.4.3.270-276.1976.
- Stucki G, Jackson TA, Noonan MJ. 1984. Isolation and characterisation of Serratia strains pathogenic for larvae of the New Zealand grass grub Costelytra zealandica. NZ J Sci. 27(3):255–260. http://pascal-francis.inist.fr/vibad/index.php?action=getRecordDetail&idt=8958985.
- Taghavi S, Garafola C, Monchy S, Newman L, Hoffman A, Weyens N, Barac T, Vangronsveld J, van der Lelie D. 2009. Genome survey and characterization of endophytic bacteria exhibiting a beneficial effect on growth and development of poplar trees. Appl Environ Microbiol. 75(3):748–757. doi:10.1128/AEM.02239-08.
- Thomson NA, Miln AJ, Kain WM. 1979. Biology of manuka beetle in Taranaki. Proc NZ Weed Pest Contr Conf. 32:80–85. doi:10.30843/nzpp.1979.32.10740.
- Townsend R, Dunbar E, Jackson TA. 2013. Grass grub distribution on the upper west coast defined by soil sampling and pheromone trapping. NZ Plant Pro Soc. 66:376. doi:10.30843/nzpp.2013.66.5681.
- Townsend R, Dunbar E, Jackson TA. 2018. Flight behaviour of the mānuka chafers, Pyronota festiva (Fabricius) and Pyronota setosa (Given) (Coleoptera: Melonlonthinae), on the flipped soils of Cape Foulwind on the West Coast of New Zealand. NZ Plant Pro Soc. 71:25–261. doi:10.30843/nzpp.2018.71.175.
- Trought TET, Jackson TA, French RA. 1982. Incidence and transmission of a disease of grass grub (Costelytra zealandica) in Canterbury. NZ J Experim Agri. 10(1):79–82. doi:10.1080/03015521.1982.10427847.
- Vaughan AL, Altermann E, Glare TR, Hurst MRH. 2022. Genome sequence of the entomopathogenic Serratia entomophila isolate 626 and characterisation of the species specific itaconate degradation pathway. BMC. 23:728. doi:10.1186/s12864-022-08938-2.
- Villamizar LF, Barrera G, Hurst MRH, Glare TR. 2021. Characterization of a new strain of Metarhizium novozealandicum with potential to be developed as a biopesticide. Mycology. 12(4):261–278. doi:10.1080/21501203.2021.1935359.
- Williams DJ, Grimont PAD, Cazares A, Grimont F, Ageron E, Pettigrew KA, Cazares D, Njamkepo E, Weill F-X, Heinz E, et al. 2022. The genus Serratia revisited by genomics. Nat Commun. 13:5195. doi:10.1038/s41467-022-32929-2.
- Wilson CJ, Mahanty HK, Jackson TA. 1992. Adhesion of bacteria (Serratia spp.) to the foregut of grass grub (Costelytra zealandica (White)) larvae and its relationship to the development of amber disease. Biocon Sci Technol. 2(1):59–64. doi:10.1080/09583159209355218.
- Wu JY, Zhao FQ, Bai J, Deng G, Qin S, Bao QY. 2007. Adaptive evolution of cry genes in Bacillus thuringiensis: implications for their specificity determination. Geno Proteo Bioinfo. 5:102–110. doi:10.1016/S1672-0229(07)60020-5.
- Zeigler R. 2013. The family Paenibacillaceae. Bacillus genetic stock center catalog of strains part 5. BCSG. p. 33. https://bgsc.org/_catalogs/Catpart5.pdf.
- Zhang H, Jackson TA. 2008. Autochthonous bacterial flora indicated by PCR-DGGE of 16S rRNA gene fragments from the alimentary tract of Costelytra zealandica (Coleoptera: Scarabaeidae). J App Micro. 105(5):1277–1285. doi:10.1111/j.1365-2672.2008.03867.x.
- Zydenbos SM, Townsend RJ, Lane PMS, Mansfield S, O’Callaghan M, van Koten C, Jackson TA. 2016. Effect of Serratia entomophila and diazinon applied with seed against grass grub populations on the North Island volcanic plateau. NZ Plant Pro Soc. 69:86–93. doi:10.30843/nzpp.2016.69.5919.

