ABSTRACT
Allopolyploids arise when two or more species hybridise to form an entirely new species with a duplicated genome. Although initially met with an array of potentially catastrophic challenges triggered by the combination of two diverged parental subgenomes within a single cell, countless allopolyploids worldwide demonstrate exceptional biological resilience by not only living under these unique circumstances, but thriving. The archipelago of Aotearoa New Zealand is home to an unexpectedly large number of allopolyploid species, both indigenous and introduced. Here, we review the prevalence and importance of these species from a local perspective. The benefits of allopolyploid species permeate multiple facets of life in New Zealand, from pastoral health and arable crop yield, to long-established viticulture and brewery practices, to the intrinsic nature of the land through the presence of diverse native allopolyploid flora. Consequently, the motivation behind the pursuit of New Zealand’s allopolyploid research extends beyond improvement of the global knowledgebase and also aims to drive tangible economic and cultural impacts on the country and the lives of its people.
Introduction
Polyploidy, the heritable phenomenon that generates species with increased chromosomal content, is a major force in evolution (Ren et al. Citation2018). It is widely accepted that polyploidy encompasses a continuum spanning its two primary classifications, autopolyploidy and allopolyploidy (Tayalé and Parisod Citation2013; Spoelhof et al. Citation2017). Autopolyploids, the prefix ‘auto-’ meaning ‘same’, are generated from whole genome duplication within species. Conversely, allopolyploids (‘allo-’, meaning ‘different’) originate from the hybridisation of two or more different species (Comai Citation2005). This continuum of polyploidy accounts for intermediates whose chromosomes exhibit both homologous (as in autopolyploids) and homeologous (as in allopolyploids) pairing during cell division. Intermediacy in polyploids was described in the seminal works by Stebbins (Citation1947, Citation1950), who later asserted the inefficiency of selection in duplicated genomes, deeming polyploids to be evolutionary dead-ends (Stebbins Citation1971). Stebbins’ view, the ‘dead-end hypothesis’, has seen increasing opposition through subsequent discoveries of recurrent polyploidy (Soltis and Soltis Citation1999), substantial dynamism within polyploid genomes (Scannell et al. Citation2006; Flagel and Wendel Citation2009; Parisod et al. Citation2009; Chester et al. Citation2012; Wang et al. Citation2015; Wang et al. Citation2018) and the presence of an ancestral polyploidy event at the base of the major land plant radiation (Jiao et al. Citation2011).
The genetic consequences of allopolyploidy
Allopolyploidy appears almost as an antithesis of conventional Darwinian evolution, where new species evolve from a single common ancestor, typically conceptualised as a bifurcating tree (Darwin Citation1859). In allopolyploidy, multiple ancestral species instead hybridise to form a single new species. Allopolyploid formation has been shown to be accompanied by extensive structural and biochemical changes to the genome, which have been described as ‘genome shock’ (McClintock Citation1984). Indeed, this phrase aptly captures the essence of this potentially catastrophic array of structural and biochemical changes that can occur, near-instantaneously, from ploidy elevation and hybridisation within the nascent allopolyploid genome. The suite of potential biological responses, both rapid and longer term, employed by the allopolyploid in response to genome shock have many potential manifestations, including genomic restructuring (Lim et al. Citation2008; Wu et al. Citation2015; Qin et al. Citation2016; Wang et al. Citation2018) and gene loss (Scannell et al. Citation2006; Gordon et al. Citation2009; Vallejo-Marín et al. Citation2015). Although the impact of allopolyploidy is most often viewed through the lens of nuclear genomic studies, organellar genomes (Sloan et al. Citation2018), small RNAs (Ha et al. Citation2009; Jiao et al. Citation2018) and the gene expression profiles of allopolyploid species (Yoo et al. Citation2014; Jung et al. Citation2015) are also affected. Extensive changes in allopolyploid gene expression (‘transcriptome shock’) are driven by the mixing of two dissimilar and poorly co-adapted sets of transcription factors and chromatin signatures (Cox et al. Citation2014).
The relative influences of hybridisation and ploidy elevation in driving post-allopolyploidisation genetic changes are highly debated, with these causal factors exhibiting lineage-specific differences. In allopolyploid Fragaria (strawberry) species, DNA methylation changes appear to be predominantly driven by genome doubling, whereas both genome doubling and hybridisation appear to impact gene expression changes (Wang et al. Citation2016). In contrast, comparative analysis of Arabidopsis auto- and allopolyploids has suggested that gene regulation is more influenced by hybridisation (Wang et al. Citation2006). A further study of synthetic allopolyploids obtained from Chrysanthemum nankingense (chrysanthemum) x Tanacetum vulgare (tansy) hybrids proposed that it is the interaction between the two processes that is important (Qi et al. Citation2018).
Nascent polyploids must adapt to an instantaneous increase in genomic material, with potential downstream consequences for cell regulatory functions. Such consequences are typically generated through non-proportional cellular expansion (Melaragno et al. Citation1993; Robinson et al. Citation2018), producing a stoichiometric imbalance between chromatin components and envelope proteins (Corredor et al. Citation2005). This scaling relationship may be further observed at the organismal level, particularly in polyploid plants (the gigas effect) (Sattler et al. Citation2016; Robinson et al. Citation2018), or there may be a compensatory reduction in cell number that lessens the impact of increased cell size on organ size changes (Tsukaya Citation2008; Del Pozo and Ramirez-Parra Citation2014; Czesnick and Lenhard Citation2015). Diploidisation, the process by which a polyploid genome returns to a state of diploidy, commonly follows ploidy elevation, suggesting that the challenges associated with whole genome duplication are often too complex to allow the establishment of the nascent polyploid species (Wolfe Citation2001; Leitch and Bennett Citation2004). Alternatively, diploidisation may be triggered by the unavailability of any required ecological drivers, such as novel niche availability (Baduel et al. Citation2018). As a result, some presumed classical diploid lineages are in fact paleo-polyploids, having experienced single or multiple rounds of polyploidisation and diploidisation in their evolutionary history (Wolfe and Shields Citation1997; Wolfe Citation2001; Ozkan and Feldman Citation2009; Qiao et al. Citation2019).
As well as genomic changes, allopolyploidy can also yield novel phenotypes, largely through the diversity introduced by hybridisation (Hedrick Citation2013). Allopolyploids may be morphologically intermediate between both parents due to homeolog (parental gene copy) co-dominance at a given locus (Szymura and Farana Citation1978). Alternatively, hybrids may show a phenotypic similarity to one parent as a result of subgenome dominance or genomic imprinting (Heslop-Harrison Citation1990; Edger et al. Citation2017; Bird et al. Citation2018), or they may exhibit a phenotype beyond the range of either parental species, generated through transgressive expression (Rieseberg et al. Citation1999; López-Caamal and Tovar-Sánchez Citation2014). It is possible that some phenotypic changes observed in allopolyploids result from genome doubling, rather than hybridisation, with the multiple gene copies produced through duplication able to perform different functions. Support for this hypothesis is also provided by the phenotypic differences observed between autopolyploids, whose origin does not involve hybridisation, and their diploid ancestors (Segraves and Thompson Citation1999).
Advantages of allopolyploidy
The persistence of allopolyploidy across Eukarya (Yoo et al. Citation2013; Cox et al. Citation2014; Sehrish et al. Citation2014; Session et al. Citation2016; Matos et al. Citation2019) suggests that successful adaptation to all of these challenges may grant the allopolyploid advantages previously unavailable to its parent lineages. Their doubled and hybrid genomes can facilitate intergenomic heterosis (hybrid vigour); a phenotypic consequence where hybrid species demonstrate increased biological fitness when compared with either parental line (Baranwal et al. Citation2012; Fujimoto et al. Citation2018). At a molecular level, allopolyploid hybrid vigour may manifest as a buffering effect against deleterious recessive mutations (Gu et al. Citation2003), novel gene function innovation through neo- or subfunctionalisation of genes (Adams and Wendel Citation2005), and the evolution of complementary parental homeologs at a given genetic locus (Paterson Citation2005). Heterosis and transgressive phenotypes can enable some allopolyploids to outcompete their parents within the same ecological niche, or to colonise niches that are more extreme than those of either parental species. Thus, allopolyploidy is often associated with invasiveness (Ainouche et al. Citation2008; Kim et al. Citation2008; Pandit et al. Citation2011).
Allopolyploidy in New Zealand
New Zealand allopolyploids include introduced species; both those that underwent allopolyploidy prior to their arrival and those that were allopolyploids upon introduction, as well as an array of native and endemic allopolyploid taxa (). Allopolyploid species play an important role in the country’s industries and economy, as well as in the mauri, or ‘vital essence’, of the land. Studies of New Zealand native allopolyploids offer complementary information and a unique perspective to global allopolyploid research, while improving our understanding of the evolution of a unique biota. Here, we discuss the importance and prevalence of allopolyploidy in Aotearoa New Zealand, in the context of the outcomes, timings and locations of their formation.
When Māori arrived in New Zealand from tropical Polynesia around AD 1250, they brought with them a number of tree and root crops (Leach and Stowe Citation2005). Only six of these species, whose cultivation was mostly marginal in New Zealand’s temperate climate, are known to have survived into European times: aute (paper mulberry, Broussonetia papyrifera), hue (bottle gourd, Lagenaria siceraria), kūmara (Ipomoea batatas), taro (Colocasia esulenta), tī pore (Corydyline fruticosa) and uwhi (yam, Dioscorea alata). Polyploidy is inferred in the origins of three of these species – kūmara (Roullier et al. Citation2013), tī pore (Hinkle Citation2004) and uwhi (Nemorin et al. Citation2012) – but research to date indicates that these origins lie in autopolyploidy rather than allopolyploidy.
Table 1. Prominent New Zealand allopolyploid taxa, with origin and commercial significance.
In contrast, post-colonial agriculture, which forms the backbone of the New Zealand economy today (Brooking Citation2006; Peden Citation2008), relies heavily on allopolyploids (). Most introduced allopolyploids formed prior to their introduction to New Zealand, but there are also examples of allopolyploidy events occurring post-introduction.
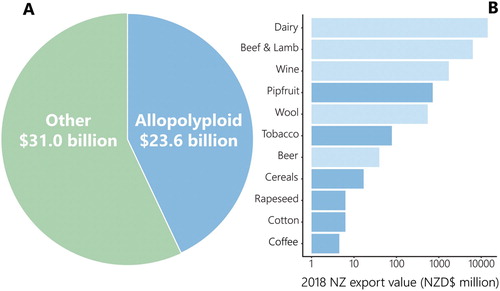
Figure 1. Economics of New Zealand allopolyploids. (A) Pie chart of New Zealand’s total exports in 2018, separated into goods that derive directly or indirectly from systems that are reliant on allopolyploid species (blue) and other goods (green). Data were obtained using the New Zealand Trade Dashboard (Stats NZ Citation2019; accessed 24 June 2019) for the categories ‘exports’, ‘goods’ and ‘2018’. (B) Breakdown of allopolyploid export goods based on their individual contributions to the New Zealand economy in 2018 (NZ dollars, millions). Goods that are directly allopolyploid are represented by dark blue bars. Goods that derive indirectly from allopolyploid species are represented by pale blue bars. The x-axis is a log (base 10) scale.
Allopolyploidisation pre-introduction
Agricultural exports exceed NZD$25 billion per annum, most of which are products derived from pasture-grazing livestock (Ministry of Business, Innovation & Employment Citation2018). The 2017 census quantified national sheep, dairy cattle and beef cattle numbers at 27.5, 6.5 and 3.6 million, respectively (Stats NZ Citation2018). The human population of New Zealand is approaching 5 million. For the agricultural sector to maintain these consistently high economic contributions, it is heavily reliant on pastoral quality and condition. Endophyte species of the genus Epichloë live as obligate symbionts of the cool-season grasses found in New Zealand pastures, particularly perennial ryegrass (Lolium perenne). An unexpectedly large number of Epichloë species have arisen via allopolyploidisation, with the genus subsequently becoming an emerging model system for studying fungal allopolyploidy (Schardl et al. Citation1994; Schardl et al. Citation2013; Cox et al. Citation2014; Campbell et al. Citation2017). Although central to New Zealand’s agronomy, Epichloë species seldom form symbioses with native grasses (Rolston et al. Citation2002), although endemic Epichloë species are often allopolyploid (Leuchtmann et al. Citation2019). Agricultural strains were first introduced via seed brought by British immigrants during the 1800s (Stewart Citation2006). The coevolution between Epichloë endophytes and their hosts is thought to drive the specialisation and mutualistic cooperation of these symbioses (Leuchtmann et al. Citation1997; Saikkonen et al. Citation2004; Saikkonen et al. Citation2016). Able to persist asymptomatically within their host tissue, Epichloë fungi produce beneficial secondary metabolites that act as natural insecticides and enhance the survival of their grass host, in addition to improving the vegetative growth and drought tolerance of the pasture (Song and Nan Citation2015; Saikkonen et al. Citation2016).
Epichloë species are estimated to contribute NZD$200 million to the New Zealand economy per annum (Johnson et al. Citation2013). Given that one endophyte strain, E. festucae var. lolii AR37, introduced commercially at the 2006 Fieldays, is estimated to have taken NZD$12 million in development costs (Ministry of Business, Innovation & Employment Citation2018), this return has proved an immensely rewarding investment for New Zealand and its future. Different Epichloë strains make different compounds that protect their grass host from insect herbivory, with allopolyploid species often producing the most diverse array of compounds. While AR37 is not an allopolyploid, other allopolyploid strains such as E. hybrida Lp1 have been commercialised in other agricultural settings, and due to their diverse mechanisms of anti-insect protection, many contenders for third-generation commercial products are also allopolyploids. Beyond these economic incentives, Epichloë research also contributes fundamental information, such as the impacts of allopolyploidy on modulating fungal gene expression, to the growing global knowledgebase on allopolyploid species (Cox et al. Citation2014).
Epichloë endophytes are not the only allopolyploid species central to the functioning of the agricultural sector. White clover (Trifolium repens) is a key pastoral plant, owing to its quality forage material and nitrogen-fixing abilities (Charlton Citation2008). White clover emerged following an allopolyploidisation event between two European clover species confined to markedly different coastal and alpine habitats. Genetic analyses suggest that their unlikely co-habitation was driven by the reconfigured European landscape during the last glacial period (Griffiths et al. Citation2019). New Zealand’s history of white clover research extends back some 80 years, incentivised by its advantages for pastoral agriculture. Nitrogen fixation by white clover has enabled pasture establishment in regions with significant mineral deficits, including the volcanic plateau and Northland gumlands (Brock et al. Citation1989). Moreover, a higher pastoral clover content has been positively correlated with dairy cow milk yields (Harris et al. Citation1998). White clover is a remarkable example of allopolyploidy-facilitated niche expansion that produced a ubiquitous temperate forage crop able to outcompete its two highly-specialised parental species, with surprising benefits for the modern pastoral economy. Continued research in New Zealand on the interactions between the two parental subgenomes will enable the selection of superior white clover strains for use in pastoral breeding programmes (Griffiths et al. Citation2019).
Since the 1850s, pastoralism has dominated agricultural production in New Zealand, but crop production still constitutes an important source of economic gain in the agricultural sector (Peden Citation2008). New Zealand’s major arable crops include a number of allopolyploid species formed prior to their introduction (). Cotton crops (Gossypium species) are a striking example of hybrid vigour through the evolution of complementary parental homeologs. Modern-day cotton cultivars are naturally-formed allotetraploids whose extinct diploid parental species, most closely related to extant G. arboreum and G. raimondii (Wendel et al. Citation1995), hybridised 1–2 million years ago (Hu et al. Citation2015). Notably, G. raimondii possesses a phenotype devoid of any spinnable fibre qualities (Hu et al. Citation2015). The natural union of complementary homeologs from these two divergent genomes in a common allopolyploid nucleus, in combination with intense human-mediated artificial selection, has seen the development of cotton species whose maximum yield, and fibre strength, length and fineness consistently surpass those of their parental species (Paterson Citation2005). New Zealand’s cotton exports, a sector not widely recognised, exceeded NZD$6 million in 2018 (B), mainly in the form of woven cotton fabrics, as opposed to combed or carded raw cotton fibres (Stats NZ Citation2019).
As yet another example of allopolyploidy in agriculture, many areas of New Zealand, including Nelson, Hawke’s Bay and Central Otago, provide an ideal climate for the cultivation of apples (Malus species); a purportedly allopolyploid crop (Chevreau and Laurens Citation1987; Tatum et al. Citation2005) that has been grown on New Zealand soil since European settlement and exported since the 1880s. Several new apple varieties have been developed in New Zealand, including Royal Gala, Jazz and Lemonade. The sequencing of the domestic apple genome in 2010 gave unprecedented insight into the evolutionary history of the Malus genus, postulating a polyploidy event that occurred over 50 million years ago in the ancestral lineage from which another pipfruit, pears (Pyrus species), have also originated. This duplication may have enabled the expansion of key gene families, such as those implicated in carbohydrate metabolism, a gene family over-represented in apples (Velasco et al. Citation2010), and those responsible for the commonly observed red fruit-skin pigmentation (Chagné et al. Citation2013). However, contrary to earlier studies, more recent genomic analysis is favouring the view that apples may have originated through autopolyploidy (Velasco et al. Citation2010). The evolutionary history of pears is also unclear, with some studies supporting an allopolyploid origin (Evans et al. Citation2008), while others support the autopolyploidy hypothesis (Li et al. Citation2019). The latter study drew their conclusions from the observed unbiased subgenome evolution in the pear genome; a common trend among paleo-autopolyploids, but one that has also been observed in some paleo-allopolyploid lineages (Sun et al. Citation2017; Li et al. Citation2019).
Introduced allopolyploid taxa of importance to the New Zealand economy are also prevalent well beyond the agricultural and horticultural sectors. Saccharomyces, the fungal genus of the model organism S. cerevisiae (brewer’s and baker’s yeast), is instrumental in brewery practices worldwide. Saccharomyces cerevisiae is the descendent of an ancient allopolyploidisation event, although genetic analyses have revealed extensive diploidisation, resulting in only minor retention of duplicated gene copies, most commonly those associated with beneficial novel properties (Wolfe Citation2001; Pfliegler et al. Citation2012; Marcet-Houben and Gabaldón Citation2015; Wolfe Citation2015). Notably, S. cerevisiae, an ale (top fermenting) yeast, is also a parent of the allopolyploid lager (bottom fermenting) yeast, S. pastorianus (Casaregola et al. Citation2001; Lodolo et al. Citation2008). However, it is the genomic contribution of its other parental species, S. eubayanus, that granted S. pastorianus the specific sugar and sulphite metabolism changes necessary for its lager-brewing domestication (Libkind et al. Citation2011).
The establishment of New Zealand vineyards also began in the early 1800s. Domesticated allopolyploid S. cerevisiae of course plays a central role in wine making too, dominating the initial (Mangado et al. Citation2018) and late (Bagheri et al. Citation2017) stages of fermentation due to its high sugar processing rates and ethanol tolerance. Recent evidence suggests that S. cerevisiae not only dominates this ecosystem, but also directly influences the other constituent species (Bagheri et al. Citation2017). Seemingly in parallel with the allopolyploidy observed in brewing yeasts, strains of Dekkera bruxellensis, a prominent industrial wine fermentation contaminant, also appear to be allopolyploids (Borneman et al. Citation2014). The separation of the Saccharomyces and Dekkera lineages occurred at least 200 million years ago, preceding the ancient allopolyploidisation event in the S. cerevisiae lineage (Schifferdecker et al. Citation2014). Perhaps this genotypic convergence has been driven by the biological pressure from an evolutionary arms race between these two fermentative yeasts. A purported link exists between the ploidy level of Saccharomyces species and culture productivity (Albertin and Marullo Citation2012), and is a plausible explanation for the strong presence of allopolyploids among brewing yeasts, particularly when considered in conjunction with hybrid vigour. Subsequently, it is in the interest of New Zealand brewers to explore the existence of novel local allopolyploid strains that may provide a unique new edge to New Zealand’s longstanding brewing and viticultural practices.
Allopolyploidisation post-introduction
Allopolyploid taxa whose hybridisation occurred following their introduction to New Zealand are less common than the examples above where allopolyploidisation preceded their arrival in New Zealand. One case of hybridisation subsequent to arrival, which has benefited the brewing industry, is triploid hops (Humulus lupulus). The New Zealand brewing industry pioneered the development of triploid hops, first releasing a commercial variety in 1972 in response to the growing global demand for a seedless phenotype. Today, 24 New Zealand-developed triploid cultivars are available (New Zealand Hops Ltd. Citation2018), distinguishable beyond their seedlessness by their superior growth and lushness relative to diploid cultivars (Trojak-Goluch and Skomra Citation2018). Although strictly autopolyploids, triploid hops are formed by controlled crosses between genetically extremely distinct cultivars; most often between a tetraploid female and diploid male, and thus nonetheless contain diverse parental genomes (Beatson and Brewer Citation1994; Beatson et al. Citation2003).
New Zealand’s large population of introduced flora provides ample opportunity for naturally-occurring allopolyploidy events among taxa following their arrival. Given the purported role of polyploidy (Pandit et al. Citation2011; te Beest et al. Citation2012; Baduel et al. Citation2018) and hybridisation (Ellstrand and Schierenbeck Citation2000; Hovick and Whitney Citation2014; Gaskin Citation2016) in plant invasions, it will be important to remain vigilant to such occurrences. Certainly, allopolyploidy appears to have been central to the invasiveness of the aggressive hawkweed (Pilosella officinarum), which was accidentally introduced over 100 years ago and has since become the bane of New Zealand’s high country (Morgan-Richards et al. Citation2004; Morgan-Richards et al. Citation2009). Data suggests that the hybridisation of P. officinarum with a related species (likely Hieracium praealtum) has occurred at least three times within New Zealand, generating plants that appear phenotypically similar to P. officinarum, but that may act as conduits for increased gene flow between the species and possess the potential to colonise from single seeds, an ability likely to facilitate the rapid spread of an invasive species (Morgan-Richards et al. Citation2004; Trewick et al. Citation2004). In contrast to the direct genetic manipulation of triploid hops, allopolyploid hawkweed appears to have arisen as a consequence of human-induced habitat disturbance (Morgan-Richards et al. Citation2009).
Native flora and fauna
Introduced allopolyploid species have become an integral part of the country: for its industries, its economy and its food-chain. Many of these species are relatively well studied, due to their analogous roles on a global scale and their economic importance (). However, New Zealand native allopolyploid species are significantly less well understood than their non-native counterparts. This knowledge gap belies the importance of native allopolyploids to the distinctiveness of the New Zealand biota. Concordant with the global trend (Jiao et al. Citation2011; Barker et al. Citation2016), a wealth of native allopolyploid diversity is found in New Zealand’s flora, with documented examples across mosses, ferns and angiosperms (flowering plants). Allopolyploidy is possibly largely absent from New Zealand native fauna, even for species with otherwise unusual reproductive strategies (Morgan-Richards et al. Citation2019). The sole case known to us is an ancient hybridisation inferred in the formation of the mosaic allotriploid, parthenogenetic Acanthoxyla stick insects (Buckley et al. Citation2008; Myers et al. Citation2013) (). Triploidy has been documented in the New Zealand freshwater snail Potamopyrgus antipodarium (Soper et al. Citation2013), although its origin appears to lie in autopolyploidy (Neiman et al. Citation2011).
Mosses
Mosses are considered to be one of the earliest forms of land plant that evolved in adaptation to the higher CO2 levels present from the early Paleozoic Era (Shaw et al. Citation2011). New Zealand is home to more than 500 moss species, approximately 20% of which are considered endemic (Glenny et al. Citation2011). Although mosses are haploid-dominant organisms (Graham and Wilcox Citation2000), researchers have identified two species of the New Zealand native Sphagnum mosses (peat moss) that lack haploid gametophytes (Karlin et al. Citation2009; Karlin and Smouse Citation2017). Sphagnum australe and S. falcatulum are found throughout the South Island only as allodiploid or allotriploid cytotypes, and it further appears that the allotriploids are the predominant plant form, perhaps due to a competitive advantage over their allodiploid counterparts that allows them to inhabit a broader ecological niche. Hybridisation and polyploidy are relatively common within the Sphagnum genus (Ricca et al. Citation2008; Karlin et al. Citation2014), as is interploidal hybridisation (i.e. hybridisation between different ploidy levels) relative to angiosperms (Flatberg et al. Citation2006; Karlin et al. Citation2009; Meleshko et al. Citation2018). For S. australe and S. falcatulum allotriploids, one of the parental species is their respective allodiploid species (or ancestor thereof). Both taxa also display intersectional allopolyploidy, with S. australe appearing to have parental species from the sections Sphagnum and Rigida, and S. falcatulum from the sections Subsecunda and Cuspidata (Karlin et al. Citation2009).
Ferns
Ferns are another prominent feature of the damp understory of forested areas throughout New Zealand; the number of distinct species is unusually high for a temperate country (approximately 200, with 44% endemicity) (Schönberger et al. Citation2018). Among vascular plants globally, ferns demonstrate the highest frequency of polyploid species, with polyploidy implicated in up to 31% of speciation events (Wood et al. Citation2009). Consequently, ferns have long been used as a model system for studying polyploidy and its genetic repercussions (DeMaggio et al. Citation1971). The genus Asplenium (spleenworts) contains more than 700 species worldwide. Its Austral group, a polyploid complex centred in New Zealand (Brownsey Citation1977; Shepherd, Holland, et al. Citation2008), is unique when compared with European and North American groups of the same genus due to its absence of diploid organisms (Dawson et al. Citation2000; Perrie and Brownsey Citation2005). Seven tetraploid and eight octoploid Asplenium species are found in New Zealand, with allopolyploidy implicated in the origin of seven of the octoploid species (Shepherd, Perrie, et al. Citation2008), mirroring the high frequency of allopolyploidy found in Asplenium worldwide (Lovis Citation1978; Reichstein Citation1981; Wagner et al. Citation1993; Schneider et al. Citation2017). Interestingly, one of the tetraploids is thought to be the parent of five of the octoploids, suggesting that this single species has played a central role in the evolution of this fern complex (Shepherd, Perrie, et al. Citation2008). Despite extensive sympatry and having the same parental species, the allopolyploids A. cimmeriorum and A. gracillimum have each evolved independently at least twice, with the four lineages reproductively isolated (Perrie et al. Citation2010), thus emphasising the importance of allopolyploidy in the generation of biological diversity. It is possible that the octoploids A. shuttleworthianum and A. northlandicum have extinct parental species; nuclear sequences of the LFY gene obtained from the allopolyploids could not be identified among the tetraploid taxa analysed by Shepherd, Perrie, et al. (Citation2008). However, it is also possible that these sequences were inherited from unsampled Pacific species.
The genus Polysticum (shield ferns) contains an allopolyploid complex located in New Zealand, and provided the first evidence-based demonstration of allopolyploidy in the New Zealand fern flora (Perrie et al. Citation2003). Initially considered to be a single species with very high levels of morphological variability (Allan Citation1961; Brownsey and Smith-Dodsworth Citation1989), P. richardii is now recognised as an allopolyploid complex containing four unique evolutionary lineages: two allooctoploid subspecies and two purportedly-parental tetraploid species. Allooctoploid P. neozelandicum is widely distributed throughout New Zealand, including on several outer islands. However, the individual distributions of its two subspecies do not overlap. Notably, allooctoploid P. neozelandicum subsp. zerophyllum has a wider geographical distribution than either of its tetraploid parental species, despite possessing relatively lower intra-lineage genetic variability (Perrie et al. Citation2003). It is possible that the New Zealand Polystichum complex represents a further example of allopolyploidy-mediated niche expansion and hybrid vigour. However, a wider study of the geographic ranges of all New Zealand ferns and lycophytes has found no obvious correlation between range sizes and polyploidy (Mountier et al. Citation2018).
Angiosperms
Angiosperms are the largest group of vascular plants, numbering over 300,000 species worldwide. Phylogenomic evidence has implicated an ancient whole genome duplication event in the ancestry of all extant angiosperm lineages (Jiao et al. Citation2011). Allopolyploidy, specifically, has been instrumental in the evolution of many of these (Osabe et al. Citation2012; Lyu Citation2016), and importantly, the link between New Zealand and allopolyploid angiosperms extends well beyond those species that make major contributions to the economy via crop production. Among New Zealand native angiosperms, there is an excess of species with even haploid numbers (i.e. the number of chromosomes within a single set), and an excess of those with haploid numbers greater than 10–14. These two karyotypic features are strong indicators of polyploid ancestry and substantiate the key role of polyploidy in the evolution of the New Zealand flora (Murray and de Lange Citation2011).
Perhaps the best-studied example of native allopolyploidy is Pachycladon (New Zealand rockcress), a genus in the Brassicaceae that is related to the model organism Arabidopsis thaliana. The number of species in the Pachycladon genus increased from one to eleven in the last million years following an allopolyploidisation event during the Pleistocene (Joly et al. Citation2009; Mandáková et al. Citation2010). Today, ten Pachycladon species are endemic to the South Island, and exhibit considerable morphological and habitat diversity in mainly alpine environments. Phylogenetic modelling and molecular studies have provided strong evidence for an adaptive radiation in the New Zealand group, despite the absence of clear phenotypic adaptations (Joly et al. Citation2014), with allopolyploidy and subsequent genomic restructuring within the founding species being the purported driving force (Joly et al. Citation2009; Mandáková et al. Citation2010). An adaptive radiation in Pachycladon is an impressive example of hybridisation between significantly diverged parental species. The level of divergence between the parents prior to their hybridisation is thought to be greater than that of allopolyploids in Gossypium, a genus that has long boasted one of the highest known parental genetic divergences among allopolyploids (Joly et al. Citation2009).
Polyploidy is also frequent among the more than 200 species of the globally-widespread genus Plantago (Murray et al. Citation2010). Commonly known as plantains, Plantago primarily comprises small herbaceous plants and shrubs. The eleven species of Plantago in New Zealand are not monophyletic and form three distinct lineages that appear to derive from at least three relatively-recent long-distance dispersal events, likely from Australia (Tay et al. Citation2010). Cytological and molecular studies suggest that most, if not all, New Zealand polyploid Plantago are allopolyploids (Ishikawa et al. Citation2009; Murray et al. Citation2010). This feature appears to have been central to their evolutionary history and taxonomic complexity, generating six ploidy levels, from diploid to 16-ploid, and forming many sympatric species within the New Zealand group (Rahn Citation1957; Rattenbury Citation1957; Groves and Hair Citation1971; Murray et al. Citation2010; Meudt Citation2011).
Looking forward
Allopolyploid species, both native and introduced, are central to many key aspects of life in New Zealand, and thus the benefits they confer to the country and its people are vast, far-reaching and surprisingly underappreciated. Duplicated, hybrid genomes give allopolyploids access to a suite of potential benefits otherwise unavailable to their ancestral species, assuming they survive the initial genome shock. The prevalence of allopolyploid species throughout New Zealand attests not only to their resilience in overcoming these challenges, but also to their ability to thrive and oftentimes surpass the biological fitness of their parental species. In some cases, such as the allopolyploid hawkweed, this ability is clearly demonstrated through invasiveness. In others, humans have been able to control the distribution of allopolyploid species, and even manipulate their ploidy level. New Zealand demonstrates a high level of polyploidy among its vascular plants: early studies characterised polyploidy in approximately 63% of angiosperms (Hair Citation1966). This value is concordant with the globally-observed positive correlation between polyploidy and latitude (Stebbins Citation1984; Brochmann et al. Citation2004; te Beest et al. Citation2012), but contrasts with the chromosomal stasis (i.e. the absence of chromosomal change) seen in other Pacific island groups (Murray and de Lange Citation2011; Stuessy et al. Citation2014). It has been postulated that the high number of introduced plant species (approximately 50%) among the native biota has generated both the opportunities, and the pressure, for increased hybridisation within New Zealand (Morgan-Richards et al. Citation2009). New Zealand allopolyploid research would benefit from a dedicated survey of the prevalence of allopolyploid taxa, both native and introduced, in place of the current rough estimates made from the independent prevalence of hybridisation and polyploidy, and the incomplete lists of known allopolyploid cases.
Statistics New Zealand data (Citation2019) suggest that allopolyploids, either directly or indirectly, collectively contribute around NZD$23 billion to the economy per annum (A), with this value excluding the largest economic sector, tourism. The need to continue to invest in the development of superior crop strains that deliver higher yields with reduced energy, water and land area requirements is strengthened by the rapidly growing global population: by 2050, the world’s population is projected to reach 9.8 billion people; 11.2 billion by the year 2100 (United Nations Department of Public Information Citation2017). The optimisation of arable crops is imperative to feed this rapidly growing global population, and the interests of New Zealand in pursuing this research rest not only on its own food sufficiency, but also on export opportunities to the rest of the world. Beyond these critical subsistence applications, allopolyploid species play an integral role in the long-established traditions of brewing and viticulture, which also make significant contributions to the New Zealand economy (B). These practices could gain a distinctive and competitive edge in both sensory profile and culture productivity through the discovery and implementation of novel native allopolyploid yeast strains.
Questions remain about the role of allopolyploidy in the evolution of New Zealand’s indigenous flora. The vast majority of New Zealand native plant species arrived via long distance dispersal (Winkworth et al. Citation2002; Wallis and Jorge Citation2018). Were the successful colonisers polyploid when they arrived, or did polyploidy occur subsequently in New Zealand? There is evidence that both situations occurred (Murray and de Lange Citation2011), but the relative contribution of allo- versus autopolyploidy has not yet been assessed. There are also questions about the timing of allopolyploidy events. Many Northern Hemisphere allopolyploids have Pleistocene origins (Abbott and Brochmann Citation2003), with the harsh environmental conditions of glacial periods thought to increase both hybridisation (Soltis et al. Citation2004) and the production of unreduced gametes (Mable Citation2004), and leading to an increased efficiency of allopolyploid species in colonising newly deglaciated areas (Comai Citation2005). The age of many of New Zealand’s allopolyploids are unknown but Pleistocene origins have been postulated for New Zealand Asplenium (Shepherd, Holland, et al. Citation2008), Pachycladon (Joly et al. Citation2009), Lepidium (Mummenhoff et al. Citation2004) and Leptinella (Himmelreich et al. Citation2014). The creation of time-calibrated phylogenies for additional taxa has the potential to provide insight into the geological drivers of allopolyploid formation within New Zealand’s island context.
Finally, how will these allopolyploids, both of agricultural and environmental importance, adjust to a changing climate? Will increased plasticity from multiple genomes mean that allopolyploid species are more resistant to climate change? Or will allopolyploids tend to be more transient on the landscape as many early writers originally proposed? Ten years ago, Morgan-Richards et al. (Citation2009) concluded their review of genetic analyses of hybridisation in New Zealand with an acknowledgement that hybridisation is a continuing focus of evolutionary biology. Today, it feels more important than ever to prioritise local allopolyploidy research to continue to improve the crops on which the economy depends, as well as to understand the processes that have generated the unique national biota of Aotearoa New Zealand.
Acknowledgements
We would like to thank Jennifer Tate (Massey University) for her constructive feedback, David Winter (Massey University) for his help during the initial stages of figure development, Thomas Buckley (Manaaki Whenua – Landcare Research) for his advice regarding allopolyploidy among New Zealand’s native fauna, and the editor and two anonymous reviewers for their thoughtful and constructive comments.
Disclosure statement
No potential conflict of interest was reported by the authors.
ORCID
Lara D. Shepherd http://orcid.org/0000-0002-7136-0017
Murray P. Cox http://orcid.org/0000-0003-1936-0236
References
- Abbott RJ, Brochmann C. 2003. History and evolution of the Arctic flora: in the footsteps of Eric Hultén. Molecular Ecology. 12(2):299–313.
- Adams KL, Wendel JF. 2005. Novel patterns of gene expression in polyploid plants. Trends in Genetics. 21(10):539–543.
- Ainouche ML, Fortune PM, Salmon A, Parisod C, Grandbastien MA, Fukunaga K, Ricou M, Misset MT. 2008. Hybridization, polyploidy and invasion: lessons from Spartina (Poaceae). Biological Invasions. 11(5):1159–1173.
- Albertin W, Marullo P. 2012. Polyploidy in fungi: evolution after whole-genome duplication. Proceedings of the Royal Society B: Biological Sciences. 279(1738):2497–2509.
- Allan HH. 1961. Flora of New Zealand. Vol. I. Wellington: Government Printer.
- Atkinson RG, Cipriani G, Whittaker DJ, Gardner RC. 1997. The allopolyploid origin of kiwifruit, Actinidia deliciosa (Actinidiaceae). Plant Systematics and Evolution. 205(1/2):111–124.
- Baduel P, Bray S, Vallejo-Marin M, Kolář F, Yant L. 2018. The “polyploid hop”: shifting challenges and opportunities over the evolutionary lifespan of genome duplications. Frontiers in Ecology and Evolution. 6(117):117.
- Bagheri B, Bauer FF, Setati ME. 2017. The impact of Saccharomyces cerevisiae on a wine yeast consortium in natural and inoculated fermentations. Frontiers in Microbiology. 8:1988.
- Baranwal VK, Kapoor S, Zehr UB, Mikkilineni V, Tyagi AK. 2012. Heterosis: emerging ideas about hybrid vigour. Journal of Experimental Botany. 63(18):6309–6314.
- Barker MS, Arrigo N, Baniaga AE, Li Z, Levin DA. 2016. On the relative abundance of autopolyploids and allopolyploids. New Phytologist. 210:391–398.
- Beatson RA, Brewer VR. 1994. Regional trial evaluation and cultivar selection of triploid hop hybrids. New Zealand Journal of Crop and Horticultural Science. 22(1):1–6.
- Beatson RA, Ferguson AR, Weir IE, Graham LT, Ansell KA, Ding H. 2003. Flow cytometric identification of sexually derived polyploids in hop (Humulus lupulus L.) and their use in hop breeding. Euphytica. 134(2):189–194.
- Bindler G, Plieske J, Bakaher N, Gunduz I, Ivanov N, Van der Hoeven R, Ganal M, Donini P. 2011. A high density genetic map of tobacco (Nicotiana tabacum L.) obtained from large scale microsatellite marker development. Theoretical and Applied Genetics. 123(2):219–230.
- Bird KA, VanBuren R, Puzey JR, Edger PP. 2018. The causes and consequences of subgenome dominance in hybrids and recent polyploids. New Phytologist. 220(1):87–93.
- Borneman AR, Zeppel R, Chambers PJ, Curtin CD. 2014. Insights into the Dekkera bruxellensis genomic landscape: comparative genomics reveals variations in ploidy and nutrient utilisation potential amongst wine isolates. PLOS Genetics. 10(2):e1004161.
- Brochmann C, Brysting AK, Alsos IG, Borgen L, Grundt HH, Scheen A-C, Elven R. 2004. Polyploidy in Arctic plants. Biological Journal of the Linnean Society. 82(4):521–536.
- Brock JL, Caradus JR, Hay MJM. 1989. Fifty years of white clover research in New Zealand. Proceedings of the New Zealand Grassland Association. 50:25–39.
- Brooking T. 2006. Pasture, present and future – a brief history of pastoralism in New Zealand. Wellington: Ministry of Agriculture and Forestry.
- Brownsey PJ. 1977. A taxonomic revision of the New Zealand species of Asplenium. New Zealand Journal of Botany. 15(1):39–86.
- Brownsey PJ, Smith-Dodsworth JC. 1989. New Zealand ferns and allied plants. Auckland: David Bateman Ltd.
- Buckley TR, Attanayake D, Park D, Ravindran S, Jewell TR, Normark BB. 2008. Investigating hybridization in the parthenogenetic New Zealand stick insect Acanthoxyla (Phasmatodea) using single-copy nuclear loci. Molecular Phylogenetics and Evolution. 48(1):335–349.
- Campbell MA, Tapper BA, Simpson WR, Johnson RD, Mace W, Ram A, Lukito Y, Dupont P-Y, Johnson LJ, Scott DB, et al. 2017. Epichloë hybrida, sp. nov., an emerging model system for investigating fungal allopolyploidy. Mycologia. 109(5):715–729.
- Casaregola S, Nguyen HV, Lapathitis G, Kotyk A, Gaillardin C. 2001. Analysis of the constitution of the beer yeast genome by PCR, sequencing and subtelomeric sequence hybridization. International Journal of Systematic and Evolutionary Microbiology. 51(4):1607–1618.
- Chagné D, Lin-Wang K, Espley RV, Volz RK, How NM, Rouse S, Brendolise C, Carlisle CM, Kumar S, De Silva N, et al. 2013. An ancient duplication of apple MYB transcription factors is responsible for novel red fruit-flesh phenotypes. Plant Physiology. 161(1):225–239.
- Chalhoub B, Denoeud F, Liu S, Parkin IAP, Tang H, Wang X, Chiquet J, Belcram H, Tong C, Samans B, et al. 2014. Early allopolyploid evolution in the post-Neolithic Brassica napus oilseed genome. Science. 345(6199):950.
- Charlton D. 2008. Pastures – clovers. [accessed 2019 May 19]. http://www.TeAra.govt.nz/en/pastures/page-7.
- Chester M, Gallagher JP, Symonds VV, da Silva AV C, Mavrodiev EV, Leitch AR, Soltis PS, Soltis DE. 2012. Extensive chromosomal variation in a recently formed natural allopolyploid species, Tragopogon miscellus (Asteraceae). Proceedings of the National Academy of Sciences of the United States of America. 109(4):1176–1181.
- Chevreau E, Laurens F. 1987. The pattern of inheritance in apple (Malus X domestica Borkh.): further results from leaf isozyme analysis. Theoretical and Applied Genetics. 75(1):90–95.
- Comai L. 2005. The advantages and disadvantages of being polyploid. Nature Reviews Genetics. 6(11):836–846.
- Corredor E, Díez M, Shepherd K, Naranjo T. 2005. The positioning of rye homologous chromosomes added to wheat through the cell cycle in somatic cells untreated and treated with colchicine. Cytogenetic and Genome Research. 109(1–3):112–119.
- Cox MP, Dong T, Shen G, Dalvi Y, Scott DB, Ganley ARD. 2014. An interspecific fungal hybrid reveals cross-kingdom rules for allopolyploid gene expression patterns. PLOS Genetics. 10(3):e1004180.
- Czesnick H, Lenhard M. 2015. Size control in plants–lessons from leaves and flowers. Cold Spring Harbor Perspectives in Biology. 7(8):a019190.
- Darwin C. 1859. On the origin of species by means of natural selection, or, the preservation of favoured races in the struggle for life. London: John Murray.
- Dawson MI, Brownsey PJ, Lovis JD. 2000. Index of chromosome numbers of indigenous New Zealand pteridophytes. New Zealand Journal of Botany. 38(1):25–46.
- Del Pozo JC, Ramirez-Parra E. 2014. Deciphering the molecular bases for drought tolerance in Arabidopsis autotetraploids. Plant, Cell & Environment. 37:2722–2737.
- DeMaggio AE, Wetmore RH, Hannaford JE, Stetler DA, Raghavan V. 1971. Ferns as a model system for studying polyploidy and gene dosage effects. BioScience. 21(7):313–316.
- Edger PP, Smith R, McKain MR, Cooley AM, Vallejo-Marin M, Yuan Y, Bewick AJ, Ji L, Platts AE, Bowman MJ, et al. 2017. Subgenome dominance in an interspecific hybrid, synthetic allopolyploid, and a 140-year-old naturally established neo-allopolyploid monkeyflower. The Plant Cell. 29(9):2150–2167.
- Ellstrand NC, Schierenbeck KA. 2000. Hybridization as a stimulus for the evolution of invasiveness in plants? Proceedings of the National Academy of Sciences of the United States of America. 97(13):7043–7050.
- Evans KM, Govan CL, Fernández-Fernández F. 2008. A new gene for resistance to Dysaphis pyri in pear and identification of flanking microsatellite markers. Genome. 51(12):1026–1031.
- Flagel LE, Wendel JF. 2009. Gene duplication and evolutionary novelty in plants. New Phytologist. 183(3):557–564.
- Flatberg KI, Thingsgaard K, Såstad SM. 2006. Interploidal gene flow and introgression in bryophytes: Sphagnum girgensohnii × S. russowii, a case of spontaneous neotriploidy. Journal of Bryology. 28(1):27–37.
- Fujimoto R, Uezono K, Ishikura S, Osabe K, Peacock WJ, Dennis ES. 2018. Recent research on the mechanism of heterosis is important for crop and vegetable breeding systems. Breeding Science. 68(2):145–158.
- Gaskin JF. 2016. The role of hybridization in facilitating tree invasion. AoB Plants. 9(1):plw079.
- Gaut BS, d'Ennequin MLT, Peek AS, Sawkins MC. 2000. Maize as a model for the evolution of plant nuclear genomes. Proceedings of the National Academy of Sciences USA. 97(13):7008–7015.
- Glenny D, Fife AJ, Brownsey PJ, Renner MA, Braggins JE, Beever JE, Hitchmough R. 2011. Threatened and uncommon bryophytes of New Zealand (2010 revision). New Zealand Journal of Botany. 49(2):305–327.
- Gordon JL, Byrne KP, Wolfe KH. 2009. Additions, losses, and rearrangements on the evolutionary route from a reconstructed ancestor to the modern Saccharomyces cerevisiae genome. PLOS Genetics. 5(5):e1000485.
- Graham LK, Wilcox LW. 2000. The origin of alternation of generations in land plants: a focus on matrotrophy and hexose transport. Philosophical Transactions of the Royal Society of London Series B: Biological Sciences. 355(1398):757–767.
- Griffiths AG, Moraga R, Tausen M, Gupta V, Bilton TP, Campbell MA, Ashby RL, Nagy I, Khan A, Larking A, et al. 2019. Breaking free: the genomics of allopolyploidy-facilitated niche expansion in white clover. The Plant Cell. 31(7):1466–1487.
- Groves BE, Hair JB. 1971. Contributions to a chromosome atlas of the New Zealand flora: 15 miscellaneous families. New Zealand Journal of Botany. 9(4):569–575.
- Gu Z, Steinmetz LM, Gu X, Scharfe C, Davis RW, Li W-H. 2003. Role of duplicate genes in genetic robustness against null mutations. Nature. 421(6918):63–66.
- Ha M, Lu J, Tian L, Ramachandran V, Kasschau KD, Chapman EJ, Carrington JC, Chem X, Wang X-J, Chen ZJ. 2009. Small RNAs serve as a genetic buffer against genomic shock in Arabidopsis interspecific hybrids and allopolyploids. Proceedings of the National Academy of Sciences of the United States of America. 106(42):17835–17840.
- Hair JB. 1966. Biosystematics of the New Zealand flora, 1945–1964. New Zealand Journal of Botany. 4(4):559–595.
- Harris SL, Auldist MJ, Clark DA, Jansen EB. 1998. Effects of white clover content in the diet on herbage intake, milk production and milk composition of New Zealand dairy cows housed indoors. The Journal of Dairy Research. 65(3):389–400.
- Harvey AC, Fjelldal PG, Solberg MF, Hansen T, Glover KA. 2017. Ploidy elicits a whole-genome dosage effect: growth of triploid Atlantic salmon is linked to the genetic origin of the second maternal chromosome set. BMC Genetics. 18(1):34.
- Hedrick PW. 2013. Adaptive introgression in animals: examples and comparison to new mutation and standing variation as sources of adaptive variation. Molecular Ecology. 22(18):4606–4618.
- Heslop-Harrison JS. 1990. Gene expression and parental dominance in hybrid plants. Development. 108:21–28.
- Himmelreich S, Breitwieser I, Oberprieler C. 2014. Phylogenetic relationships in the extreme polyploid complex of the New Zealand genus Leptinella (Compositae: Anthemideae) based on AFLP data. Taxon. 63(4):883–898.
- Hinkle AE. 2004. The distribution of a male sterile form of ti (Cordyline fruticosa) in Polynesia: a case of human selection? Journal of the Polynesian Society. 113(3):263–290.
- Hovick SM, Whitney KD. 2014. Hybridisation is associated with increased fecundity and size in invasive taxa: meta-analytic support for the hybridisation-invasion hypothesis. Ecology Letters. 17(11):1464–1477.
- Hu G, Wendel JF, Koh J, Yoo MJ, Chen S. 2015. Gene-expression novelty in allopolyploid cotton: a proteomic perspective. Genetics. 200(1):91–104.
- Ishikawa N, Yokoyama J, Tsukaya H. 2009. Molecular evidence of reticulate evolution in the subgenus Plantago (Plantaginaceae). American Journal of Botany. 96(9):1627–1635.
- Jiao Y, Wickett NJ, Ayyampalayam S, Chanderbali AS, Landherr L, Ralph PE, Tomsho LP, Hu Y, Liang H, Soltis PS, et al. 2011. Ancestral polyploidy in seed plants and angiosperms. Nature. 473(7345):97–100.
- Jiao W, Yuan J, Jiang S, Liu Y, Wang L, Liu M, Zheng D, Ye W, Wang X, Chen ZJ. 2018. Asymmetrical changes of gene expression, small RNAs and chromatin in two resynthesized wheat allotetraploids. Plant Journal. 93(5):828–842.
- Johnson LJ, de Bonth ACM, Briggs LR, Caradus JR, Finch SC, Fleetwood DJ, Fletcher LR, Hume DE, Johnson RD, Popay AJ, et al. 2013. The exploitation of Epichloae endophytes for agricultural benefit. Fungal Diversity. 60:171–188.
- Joly S, Heenan PB, Lockhart PJ. 2009. A Pleistocene inter-tribal allopolyploidization event precedes the species radiation of Pachycladon (Brassicaceae) in New Zealand. Molecular Phylogenetics and Evolution. 51(2):365–372.
- Joly S, Heenan PB, Lockhart PJ. 2014. Species radiation by niche shifts in New Zealand’s rockcresses (Pachycladon, Brassicaceae). Systematic Biology. 63(2):192–202.
- Jung Y, Kawaura K, Kishii M, Sakuma S, Ogihara Y. 2015. Comparison of genome-wide gene expression patterns in the seedlings of nascent allohexaploid wheats produced by two combinations of hybrids. Genes and Genetic Systems. 90(2):79–88.
- Karlin EF, Boles SB, Ricca M, Temsch EM, Greilhuber J, Shaw AJ. 2009. Three-genome mosses: complex double allopolyploid origins for triploid gametophytes in Sphagnum. Molecular Ecology. 18(7):1439–1454.
- Karlin EF, Smouse PE. 2017. Allo-allo-triploid Sphagnum × falcatulum: single individuals contain most of the Holantarctic diversity for ancestrally indicative markers. Annals of Botany. 120(2):221–231.
- Karlin EF, Temsch EM, Bizuru E, Marino J, Boles SB, Devos N, Shaw AJ. 2014. Invisible in plain sight: recurrent double allopolyploidy in the African Sphagnum×planifolium (Sphagnaceae). The Bryologist. 117(2):187–201.
- Kim S-T, Sultan SE, Donoghue MJ. 2008. Allopolyploid speciation in Persicaria (Polygonaceae): insights from a low-copy nuclear region. Proceedings of the National Academy of Sciences of the United States of America. 105(34):12370–12375.
- Lashermes P, Combes M-C, Hueber Y, Severac D, Dereeper A. 2014. Genome rearrangements derived from homoeologous recombination following allopolyploidy speciation in coffee. The Plant Journal. 78(4):674–685.
- Leach H, Stowe C. 2005. Oceanic arboriculture at the margins: the case of the Karaka (Corynocarpus laevigatus) in Aotearoa. The Journal of the Polynesian Society. 114(1):7–27.
- Leitch IJ, Bennett MD. 2004. Genome downsizing in polyploid plants. Biological Journal of the Linnean Society. 82(4):651–663.
- Leuchtmann A, Schardl CL, Penny D, Chung KR, Siegel MR. 1997. Coevolution by common descent of fungal symbionts (Epichloë spp.) and grass hosts. Molecular Biology and Evolution. 14(2):133–143.
- Leuchtmann A, Young CA, Stewart AV, Simpson WR, Hume DE, Scott B. 2019. Epichloe novae-zelandiae, a new endophyte from the endemic New Zealand grass Poa matthewsii. New Zealand Journal of Botany. 1–18.
- Li Q, Qiao X, Yin H, Zhou Y, Dong H, Qi K, Li L, Zhang S. 2019. Unbiased subgenome evolution following a recent whole-genome duplication in pear (Pyrus bretschneideri Rehd.). Horticulture Research. 6(1):34.
- Libkind D, Hittinger CT, Valério E, Gonçalves C, Dover J, Johnston M, Gonçalves P, Sampaio JP. 2011. Microbe domestication and the identification of the wild genetic stock of lager-brewing yeast. Proceedings of the National Academy of Sciences of the United States of America. 108(35):14539–14544.
- Lim KY, Soltis DE, Soltis PS, Tate J, Matyasek R, Srubarova H, Kovarik A, Pires JC, Xiong Z, Leitch AR. 2008. Rapid chromosome evolution in recently formed polyploids in Tragopogon (Asteraceae). PLOS ONE. 3(10):e3353.
- Liu Q, Lin L, Zhou X, Peterson PM, Wen J. 2017. Unraveling the evolutionary dynamics of ancient and recent polyploidization events in Avena (Poaceae). Scientific Reports. 7:41944.
- Lodolo EJ, Kock JLF, Axcell BC, Brooks M. 2008. The yeast Saccharomyces cerevisiae – the main character in beer brewing. FEMS Yeast Research. 8(7):1018–1036.
- López-Caamal A, Tovar-Sánchez E. 2014. Genetic, morphological, and chemical patterns of plant hybridization. Revista Chilena de Historia Natural. 87(1):16.
- Lovis JD. 1978. Evolutionary patterns and processes in ferns. Advances in Botanical Research. 4:229–415.
- Lyu J. 2016. Crop evolution: after allopolyploidization. Nature Plants. 2:16156.
- Mable BK. 2004. ‘Why polyploidy is rarer in animals than in plants’: myths and mechanisms. Biological Journal of the Linnean Society. 82(4):453–466.
- Mandáková T, Heenan PB, Lysak MA. 2010. Island species radiation and karyotypic stasis in Pachycladon allopolyploids. BMC Evolutionary Biology. 10:367.
- Mangado A, Morales P, Gonzalez R, Tronchoni J. 2018. Evolution of a yeast with industrial background under winemaking conditions leads to diploidization and chromosomal copy number variation. Frontiers in Microbiology. 9:1816.
- Marcet-Houben M, Gabaldón T. 2015. Beyond the whole-genome duplication: phylogenetic evidence for an ancient interspecies hybridization in the baker’s yeast lineage. PLOS Biology. 13:e1002220.
- Matos I, Machado MP, Schartl M, Coelho MM. 2019. Allele-specific expression variation at different ploidy levels in Squalius alburnoides. Scientific Reports. 9(1):3688.
- McClintock B. 1984. The significance of responses of the genome to challenge. Science. 226:792–801.
- Melaragno JE, Mehrotra B, Coleman AW. 1993. Relationship between endopolyploidy and cell size in epidermal tissue of Arabidopsis. The Plant Cell. 5(11):1661–1668.
- Meleshko O, Stenøien HK, Speed JDM, Flatberg KI, Kyrkjeeide MO, Hassel K. 2018. Is interspecific gene flow and speciation in peatmosses (Sphagnum) constrained by phylogenetic relationship and life-history traits? Lindbergia. 41(1):linbg.01107.
- Meudt HM. 2011. Amplified fragment length polymorphism data reveal a history of auto- and allopolyploidy in New Zealand endemic species of Plantago (Plantaginaceae): new perspectives on a taxonomically challenging group. International Journal of Plant Sciences. 172(2):220–237.
- Ministry of Business, Innovation & Employment. 2018. Research, science and innovation system performance report: ryegrass endophytes. [accessed 2019 June 20]. https://www.mbie.govt.nz/assets/7693f53535/research-science-and-innovation-system-performance-report-2018.pdf.
- Morgan-Richards M, Langton-Myers SS, Trewick SA. 2019. Loss and gain of sexual reproduction in the same stick insect. Molecular Ecology. 28(17):3929–3941.
- Morgan-Richards M, Smissen RD, Shepherd LD, Wallis GP, Hayward JJ, Chan CH, Chambers GK, Chapman HM. 2009. A review of genetic analyses of hybridisation in New Zealand. Journal of the Royal Society of New Zealand. 39(1):15–34.
- Morgan-Richards M, Trewick SA, Chapman HM, Krahulcova A. 2004. Interspecific hybridization among Hieracium species in New Zealand: evidence from flow cytometry. Heredity. 93(1):34–42.
- Mountier CF, Case BS, Perrie L, Brownsey P, Paterson AM, Curran TJ, Buckley HL. 2018. Patterns of range size in New Zealand ferns and lycophytes. New Zealand Journal of Ecology. 42(2):248–261.
- Mummenhoff K, Linder P, Friesen N, Bowman JL, Lee J-Y, Franzke A. 2004. Molecular evidence for bicontinental hybridogenous genomic constitution in Lepidium sensu stricto (Brassicaceae) species from Australia and New Zealand. American Journal of Botany. 91(2):254–261.
- Murray BG, Datson PM, Lai ELY, Sheath KM, Cameron EK. 2004. Polyploidy, hybridization and evolution in Pratia (Campanulaceae). New Zealand Journal of Botany. 42(5):905–920.
- Murray BG, de Lange PJ. 2011. Chromosomes and evolution in New Zealand endemic angiosperms and gymnosperms. In: Bramwell D, Caujapé-Castells J, editors. The Biology of Island Floras. Cambridge: Cambridge University Press; p. 265–283.
- Murray BG, Meudt HM, Tay ML, Garnock-Jones PJ. 2010. New chromosome counts in New Zealand species of Plantago (Plantaginaceae). New Zealand Journal of Botany. 48(3–4):197–204.
- Myers SS, Trewick SA, Morgan-Richards M. 2013. Multiple lines of evidence suggest mosaic polyploidy in the hybrid parthenogenetic stick insect lineage Acanthoxyla. Insect Conservation and Diversity. 6(4):537–548.
- Neiman M, Paczesniak D, Soper DM, Baldwin AT, Hehman G. 2011. Wide variation in ploidy level and genome size in a New Zealand freshwater snail with coexisting sexual and asexual lineages. Evolution. 65(11):3202–3216.
- Nemorin A, Abraham K, David J, Arnau G. 2012. Inheritance pattern of tetraploid Dioscorea alata and evidence of double reduction using microsatellite marker segregation analysis. Molecular Breeding. 30(4):1657–1667.
- New Zealand Hops Ltd. 2018. Harvested to perfection. [accessed 2019 September 21]. https://nz.coop/harvested-to-perfection/.
- Osabe K, Kawanabe T, Sasaki T, Ishikawa R, Okazaki K, Dennis ES, Kazama T, Fujimoto R. 2012. Multiple mechanisms and challenges for the application of allopolyploidy in plants. International Journal of Molecular Sciences. 13(7):8696–8721.
- Ozkan H, Arumuganathan K, Tuna M. 2003. Nonadditive changes in genome size during allopolyploidization in the wheat (Aegilops-Triticum) group. Journal of Heredity. 94(3):260–264.
- Ozkan H, Feldman M. 2009. Rapid cytological diploidization in newly formed allopolyploids of the wheat (Aegilops-Triticum) group. Genome. 52(11):926–934.
- Pandit MK, Pocock MJO, Kunin WE. 2011. Ploidy influences rarity and invasiveness in plants. Journal of Ecology. 99(5):1108–1115.
- Parisod C, Salmon A, Zerjal T, Tenaillon M, Grandbastien MA, Ainouche M. 2009. Rapid structural and epigenetic reorganization near transposable elements in hybrid and allopolyploid genomes in Spartina. New Phytologist. 184(4):1003–1015.
- Paterson AH. 2005. Polyploidy, evolutionary opportunity, and crop adaptation. Genetica. 123(1):191–196.
- Peden R. 2008. Farming in the economy. [accessed 2019 April 2]. http://www.TeAra.govt.nz/en/farming-in-the-economy/print.
- Perrie LR, Brownsey PJ. 2005. Insights into the biogeography and polyploid evolution of New Zealand Asplenium from chloroplast DNA sequence data. American Fern Journal. 95(1):1–22.
- Perrie LR, Brownsey PJ, Lockhart PJ, Large MF. 2003. Evidence for an allopolyploid complex in New Zealand Polystichum (Dryopteridaceae). New Zealand Journal of Botany. 41(2):189–215.
- Perrie LR, Shepherd LD, De Lange PJ, Brownsey PJ. 2010. Parallel polyploid speciation: distinct sympatric gene-pools of recurrently derived allo-octoploid Asplenium ferns. Molecular Ecology. 19(14):2916–2932.
- Pfliegler WP, Antunovics Z, Sipiczki M. 2012. Double sterility barrier between Saccharomyces species and its breakdown in allopolyploid hybrids by chromosome loss. FEMS Yeast Research. 12(6):703–718.
- Qi X, Wang H, Song A, Jiang J, Chen S, Chen F. 2018. Genomic and transcriptomic alterations following intergeneric hybridization and polyploidization in the Chrysanthemum nankingense × Tanacetum vulgare hybrid and allopolyploid (Asteraceae). Horticulture Research. 5:5.
- Qiao X, Li Q, Yin H, Qi K, Li L, Wang R, Zhang S, Paterson AH. 2019. Gene duplication and evolution in recurring polyploidization-diploidization cycles in plants. Genome Biology. 20(1):38.
- Qin Q, Lai Z, Cao L, Xiao Q, Wang Y, Liu S. 2016. Rapid genomic changes in allopolyploids of Carassius auratus red var. (♀) × Megalobrama amblycephala (♂). Scientific Reports. 6:34417.
- Rahn K. 1957. Chromosome numbers in Plantago. Botanisk Tidsskrift. 53:369–378.
- Rattenbury JA. 1957. Chromosome numbers in New Zealand angiosperms. Transactions of the Royal Society of New Zealand. 84:936–938.
- Reichstein T. 1981. Hybrids in European Aspleniaceae (Pteridophyta) in Botanica Helvetica. Berichte der Schweizerischen Botanischen Gesellschaft = Bulletin de la Societe botanique suisse. 91:89–139.
- Ren R, Wang H, Guo C, Zhang N, Zeng L, Chen Y, Ma H, Qi J. 2018. Widespread whole genome duplications contribute to genome complexity and species diversity in angiosperms. Molecular Plant. 11(3):414–428.
- Ricca M, Beecher FW, Boles SB, Temsch E, Greilhuber J, Karlin EF, Shaw AJ. 2008. Cytotype variation and allopolyploidy in North American species of the Sphagnum subsecundum complex (Sphagnaceae). American Journal of Botany. 95(12):1606–1620.
- Rieseberg LH, Archer MA, Wayne RK. 1999. Transgressive segregation, adaptation and speciation. Heredity. 83(4):363–372.
- Robinson DO, Coate JE, Singh A, Hong L, Bush M, Doyle JJ, Roeder AHK. 2018. Ploidy and size at multiple scales in the Arabidopsis sepal. The Plant Cell. 30(10):2308–2329.
- Rolston MP, Stewart AV, Latch GCM, Hume DE. 2002. Endophytes in New Zealand grass seeds: occurrence and implications for conservation of grass species. New Zealand Journal of Botany. 40(3):365–372.
- Roullier C, Duputié A, Wennekes P, Benoit L, Fernández Bringas VM, Rossel G, Tay D, McKey D, Lebot V. 2013. Disentangling the origins of cultivated sweet potato (Ipomoea batatas (L.) Lam.). PLOS ONE. 8(5):e62707.
- Saikkonen K, Wäli P, Helander M, Faeth SH. 2004. Evolution of endophyte-plant symbioses. Trends in Plant Science. 9(6):275–280.
- Saikkonen K, Young CA, Helander M, Schardl CL. 2016. Endophytic Epichloë species and their grass hosts: from evolution to applications. Plant Molecular Biology. 90(6):665–675.
- Sattler MC, Carvalho CR, Clarindo WR. 2016. The polyploidy and its key role in plant breeding. Planta. 243(2):281–296.
- Scannell DR, Byrne KP, Gordon JL, Wong S, Wolfe KH. 2006. Multiple rounds of speciation associated with reciprocal gene loss in polyploid yeasts. Nature. 440(7082):341–345.
- Schardl CL, Florea S, Pan J, Nagabhyru P, Bec S, Calie PJ. 2013. The epichloae: alkaloid diversity and roles in symbiosis with grasses. Current Opinion in Plant Biology. 16(4):480–488.
- Schardl CL, Leuchtmann A, Tsai HF, Collett MA, Watt DM, Scott DB. 1994. Origin of a fungal symbiont of perennial ryegrass by interspecific hybridization of a mutualist with the ryegrass choke pathogen, Epichloë typhina. Genetics. 136(4):1307–1317.
- Schifferdecker AJ, Dashko S, Ishchuk OP, Piškur J. 2014. The wine and beer yeast Dekkera bruxellensis. Yeast. 31(9):323–332.
- Schneider H, Liu H-M, Chang Y-F, Ohlsen D, Perrie LR, Shepherd L, Kessler M, Karger DN, Hennequin S, Marquardt J, et al. 2017. Neo- and Paleopolyploidy contribute to the species diversity of Asplenium—the most species-rich genus of ferns. Journal of Systematics and Evolution. 55(4):353–364.
- Schönberger I, Wilton AD, Brownsey PJ, Perrie L, Boardman KF, Breitwieser I, Cochrane M, de Pauw B, Fife AJ, Ford KA, et al. 2018. Checklist of the New Zealand flora – ferns and lycophytes [dataset]. Lincoln: Manaaki Whenua-Landcare Research.
- Segraves KA, Thompson JN. 1999. Plant polyploidy and pollination: floral traits and insect visits to diploid and tetraploid Heuchera grossulariifolia. Evolution. 53(4):1114–1127.
- Sehrish T, Symonds VV, Soltis DE, Soltis PS, Tate JA. 2014. Gene silencing via DNA methylation in naturally occurring Tragopogon miscellus (Asteraceae) allopolyploids. BMC Genomics. 15(1):701.
- Session AM, Uno Y, Kwon T, Chapman JA, Toyoda A, Takahashi S, Fukui A, Hikosaka A, Suzuki A, Kondo M, et al. 2016. Genome evolution in the allotetraploid frog Xenopus laevis. Nature. 538:336.
- Shaw AJ, Szövényi P, Shaw B. 2011. Bryophyte diversity and evolution: windows into the early evolution of land plants. American Journal of Botany. 98(3):352–369.
- Shepherd LD, Holland BR, Perrie LR. 2008. Conflict amongst chloroplast DNA sequences obscures the phylogeny of a group of Asplenium ferns. Molecular Phylogenetics and Evolution. 48(1):176–187.
- Shepherd LD, Perrie LR, Brownsey PJ. 2008. Low-copy nuclear DNA sequences reveal a predominance of allopolyploids in a New Zealand Asplenium fern complex. Molecular Phylogenetics and Evolution. 49(1):240–248.
- Sloan DB, Warren JM, Williams AM, Wu Z, Abdel-Ghany SE, Chicco AJ, Havird JC. 2018. Cytonuclear integration and co-evolution. Nature Reviews Genetics. 19(10):635–648.
- Soltis DE, Soltis PS. 1999. Polyploidy: recurrent formation and genome evolution. Trends in Ecology & Evolution. 14(9):348–352.
- Soltis DE, Soltis PS, Tate JA. 2004. Advances in the study of polyploidy since plant speciation. New Phytologist. 161(1):173–191.
- Song H, Nan Z. 2015. Origin, divergence, and phylogeny of asexual Epichloë endophyte in Elymus species from western China. PLOS ONE. 10(5):e0127096.
- Soper DM, Neiman M, Savytskyy OP, Zolan ME, Lively CM. 2013. Spermatozoa production by triploid males in the New Zealand freshwater snail Potamopyrgus antipodarum. Biological Journal of the Linnean Society. 110(1):227–234.
- Spoelhof JP, Soltis PS, Soltis DE. 2017. Pure polyploidy: closing the gaps in autopolyploid research. Journal of Systematics and Evolution. 55(4):340–352.
- Stats NZ. 2018. Agricultural production statistics: June 2017. New Zealand Government; [accessed 2019 June 20]. https://www.stats.govt.nz/information-releases/agricultural-production-statistics-june-2017-final.
- Stats NZ. 2019. New Zealand Trade Dashboard. [accessed 2019 June 24]. https://statisticsnz.shinyapps.io/trade_dashboard/.
- Stebbins GL. 1947. Demerec M, editor. Types of polyploids: their classification and significance. Vol. 1. Academic Press. Advances in genetics.
- Stebbins GL. 1950. Variation and evolution in plants. Oxford: Oxford University Press.
- Stebbins GL. 1971. Chromosomal evolution in higher plants. London: Edward Arnold.
- Stebbins GL. 1984. Polyploidy and the distribution of the Arctic-alpine flora: new evidence and a new approach. Botanica Helvetica. 94:1–13.
- Stewart AV. 2006. Mercer CF, editor. Genetic origins of New Zealand perennial ryegrass (Lolium perenne) cultivars. New Zealand Grassland Association. ‘Breeding for success: diversity in action’ Proceedings of the 13th Australasian Plant Breeding Conference, Christchurch, New Zealand 18-21 April 2006.
- Stuessy TF, Takayama K, López-Sepúlveda P, Crawford DJ. 2014. Interpretation of patterns of genetic variation in endemic plant species of oceanic islands. Botanical Journal of the Linnean Society. 174(3):276–288.
- Sun H, Wu S, Zhang G, Jiao C, Guo S, Ren Y, Zhang J, Zhang H, Gong G, Jia Z, et al. 2017. Karyotype stability and unbiased fractionation in the paleo-allotetraploid Cucurbita genomes. Molecular Plant. 10(10):1293–1306.
- Szymura JM, Farana I. 1978. Inheritance and linkage analysis of five enzyme loci in interspecific hybrids of toadlets, genus Bombina. Biochemical Genetics. 16(3):307–319.
- Tatum TC, Stepanovic S, Biradar DP, Rayburn AL, Korban SS. 2005. Variation in nuclear DNA content in Malus species and cultivated apples. Genome. 48(5):924–930.
- Tay ML, Meudt HM, Garnock-Jones PJ, Ritchie PA. 2010. DNA sequences from three genomes reveal multiple long-distance dispersals and non-monophyly of sections in Australasian Plantago (Plantaginaceae). Australian Systematic Botany. 23(1):47–68.
- Tayalé A, Parisod C. 2013. Natural pathways to polyploidy in plants and consequences for genome reorganization. Cytogenetic and Genome Research. 140(2–4):79–96.
- te Beest M, Le Roux JJ, Richardson DM, Brysting AK, Suda J, Kubesová M, Pysek P. 2012. The more the better? The role of polyploidy in facilitating plant invasions. Annals of Botany. 109(1):19–45.
- Trewick SA, Morgan-Richards M, Chapman HM. 2004. Chloroplast DNA diversity of Hieracium pilosella (Asteraceae) introduced to New Zealand: reticulation, hybridization, and invasion. American Journal of Botany. 91(1):73–85.
- Trojak-Goluch A, Skomra U. 2018. Breeding of triploid common hop cultivars (Humulus lupulus L. Polish Journal of Agronomy. 34:3–10.
- Tsukaya H. 2008. Controlling size in multicellular organs: focus on the leaf. PLOS Biology. 6(7):e174.
- United Nations Department of Public Information. 2017. World population projected to reach 9.8 billion in 2050, and 11.2 billion in 2100 – says UN. https://www.un.org/development/desa/en/news/population/world-population-prospects-2017.html.
- Vallejo-Marín M, Buggs RJA, Cooley AM, Puzey JR. 2015. Speciation by genome duplication: repeated origins and genomic composition of the recently formed allopolyploid species Mimulus peregrinus. Evolution. 69(6):1487–1500.
- Velasco R, Zharkikh A, Affourtit J, Dhingra A, Cestaro A, Kalyanaraman A, Fontana P, Bhatnagar SK, Troggio M, Pruss D, et al. 2010. The genome of the domesticated apple (Malus × domestica Borkh. Nature Genetics. 42(10):833–839.
- Wagner WH, Moran RC, Werth CR. 1993. Flora of North America Editorial Committee, editor. Aspleniaceae. Vol. 23. Oxford University Press on Demand. Flora of North America: volume 2: pteridophytes and gymnosperms.
- Wallis GP, Jorge F. 2018. Going under down under? Lineage ages argue for extensive survival of the Oligocene marine transgression on Zealandia. Molecular Ecology. 27(22):4368–4396.
- Wang T, Liu L, Ning C, Lü Z, Jia X, Gao Z, Qiao Y. 2016. Alterations of DNA methylation and gene expression during hybridization and polyploidization in Fragaria spp. Scientia Horticulturae. 201:218–224.
- Wang J, Tian L, Lee H-S, Wei NE, Jiang H, Watson B, Madlung A, Osborn TC, Doerge RW, Comai L, et al. 2006. Genomewide nonadditive gene regulation in Arabidopsis allotetraploids. Genetics. 172(1):507–517.
- Wang M, Wang P, Lin M, Ye Z, Li G, Tu L, Shen C, Li J, Yang Q, Zhang X. 2018. Evolutionary dynamics of 3D genome architecture following polyploidization in cotton. Nature Plants. 4(2):90–97.
- Wang J, Ye LH, Liu QZ, Peng LY, Liu W, Yi XG, Wang YD, Xiao J, Xu K, Hu FZ, et al. 2015. Rapid genomic DNA changes in allotetraploid fish hybrids. Heredity. 114(6):601–609.
- Wendel JF, Schnabel A, Seelanan T. 1995. Biderectional interlocus concerted evolution following allopolyploid speciation in cotton (Gossypium). Proceedings of the National Academy of Sciences of the United States of America. 92(1):280–284.
- Winkworth R, Wagstaff S, Glenny DJ, Lockhart P. 2002. Plant dispersal N.E.W.S. from New Zealand. Trends in Ecology & Evolution. 17:514–520.
- Wolfe KH. 2001. Yesterday’s polyploids and the mystery of diploidization. Nature Reviews Genetics. 2(5):333–341.
- Wolfe KH. 2015. Origin of the yeast whole-genome duplication. PLOS Biology. 13(8):e1002221.
- Wolfe KH, Shields DC. 1997. Molecular evidence for an ancient duplication of the entire yeast genome. Nature. 387(6634):708–713.
- Wood TE, Takebayashi N, Barker MS, Mayrose I, Greenspoon PB, Rieseberg LH. 2009. The frequency of polyploid speciation in vascular plants. Proceedings of the National Academy of Sciences of the United States of America. 106(33):13875–13879.
- Wu Y, Sun Y, Shen K, Sun S, Wang J, Jiang T, Cao S, Josiah SM, Pang J, Lin X, et al. 2015. Immediate genetic and epigenetic changes in F1 hybrids parented by species with divergent genomes in the rice genus (Oryza). PLOS ONE. 10(7):e0132911.
- Yoo MJ, Liu X, Pires JC, Soltis PS, Soltis DE. 2014. Nonadditive gene expression in polyploids. Annual Review of Genetics. 48:485–517.
- Yoo MJ, Szadkowski E, Wendel JF. 2013. Homeolog expression bias and expression level dominance in allopolyploid cotton. Heredity. 110:171–180.
