Abstract
A sequence of 375 nucleotides, which included the region encoding the cleavage activation site and signal peptide of the fusion protein gene, was determined for 178 isolates of the pigeon variant strain of Newcastle disease virus (PPMV-1). These were compared with the sequences of 47 similar isolates published by GenBank, which included 30 isolates from pigeons and 17 representatives from each sublineage of avian paramyxovirus type 1. The resulting alignment was analysed phylogenetically using maximum likelihood and the results are presented as unrooted phylogenetic trees. By phylogenetic analysis all the PPMV-1 isolates except one were placed in lineage 4b (VIb). Within this lineage there was considerable genetic heterogeneity, which appears to be predominantly influenced by the date of isolation and, to a lesser extent, geographical origins of the isolates. There were two large distinguishable groups, 4bi and 4bii. The earliest isolate available, PIQPI78442, isolated in 1978 in Iraq, was situated at the node from which the two groups diverge.
Résumé
Investigations épidémiologiques et moléculaires des souches virales variantes d'APMV-1 (PPMV-1) responsables de la panzootie de 1978 à 2002 chez le pigeon
Une séquence de 375 nucléotides, qui inclut la région codant le site d'activation du clivage et le peptide signal du gène de la protéine de fusion, a été déterminée pour 178 souches variantes du virus de la maladie de Newcastle isolées chez le pigeon (PPMV-1). Ces souches ont été comparées avec les séquences de 47 souches similaires publiées dans GenBank, qui incluaient 30 souches isolées de pigeons et 17 représentants de chacun des sous lignages des paramyxovirus aviaires de type 1 (APMV-1). Le résultat des alignements a été analysé phylogénétiquement utilisant la probabilité maximale et les résultats sont présentés comme des arbres phylogénétiques sans racine. L'analyse phylogénétique a permis de placer dans le lignage 4b (VIb) toutes les souches de PPMV-1 à l'exception d'une seule. A l'intérieur de ce lignage il y a une hétérogénéité génétique considérable qui apparaît être influencée de façon prédominante par la date d'isolement et, à un moindre degré par les origines géographiques de ces souches. Il y a deux grands groupes distincts, 4bi et 4bii. La première souche disponible, PIQPI78442, isolée en Irak en 1978, est située au nœud où les deux groupes divergent.
Zusammenfassung
Eine molekular-epidemiologische Untersuchung von Isolaten der PPMV-1-Variante des APMV-1, die für die 1978-2002-Panzootie in Tauben verantwortlich waren
Eine Sequenz von 375 Nukleotiden, die die Region umfasste, die für die Cleavage-Aktivierungsstelle und das Signalpeptid des Fusionsproteingens kodiert, wurde bei 178 Isolaten der Taubenvariante des Virus der Newcastle-Krankheit (PPMV-1) bestimmt. Diese wurden mit den Sequenzen von 47 ähnlichen, von der GenBank publizierten Isolaten, 30 Isolaten aus Tauben und 17 Vertretern von jeder der Sublinien des aviären Paramyxovirus-Typ 1 (APMV-1), verglichen. Die resultierenden Gruppierungen wurden phylogenetisch anhand maximaler Ähnlichkeiten analysiert und die Ergebnisse wurden als wurzellose phylogentische Bäume dargestellt. Durch die phylogenetische Analyse wurden alle PPMV-1-Isolate bis auf eines der Linie 4b (VIb) zugeordnet. Innerhalb dieser Linie gab es eine beachtliche genetische Heterogenität, die hauptsächlich vom Isolierungsdatum und in geringerem Maße von der geographischen Herkunft der Isolate beeinflußt zu werden schien. Es ließen sich zwei große Gruppen, 4bi und 4bii, unterscheiden. Das früheste verfügbare Isolat, PIQPI78442, das 1978 im Irak isoliert worden war, liegt an dem Knotenpunkt, von welchem aus die beiden Gruppen auseinander gehen.
Resumen
Epidemiología molecular de los aislados del virus variante APMV-1 (PPMV-1) responsable de la panzootia de 1978 –2002 en palomas
Se determinó una secuencia de 375 nucleótidos, que incluía la región que codifica para el punto de activación de la escisión y el péptido señal del gen de la proteína de fusión, de 178 aislados de la cepa variante de paloma del virus de la enfermedad de Newcastle (PPMV-1). Estas secuencias fueron comparadas con secuencias de 47 asilados similares publicadas en el GenBank, que incluían 30 aislados de paloma y 17 representativos de cada uno de los sublinajes del paramixovirus aviar tipo 1 (APMV-1). El alineamiento resultante fue analizado mediante maximum likelihood y los resultados fueron presentados como árboles filogenéticos sin raíz. Mediante el análisis filogenético todos los aislados de PPMV-1 excepto uno se clasificaron dentro del linaje 4b (VIb). En este linaje hay una considerable heterogeneidad genética, que parece estar influenciada mayoritariamente por el año de aislamiento y , en menor medida, por el origen geográfico de los aislados. Se encontraron dos grandes grupos, 4bi y 4bii. El aislado disponible más antiguo, PIQPI78442, aislado en 1978 en Iraq, se situó en el nudo a partir del cual los dos grupos divergían.
Introduction
A series of outbreaks of low-morbidity, low-mortality Newcastle disease were reported in flocks of pigeons (Columba livia) across south eastern Europe and Italy during the early 1980s (Biancifiori & Fioroni, Citation1983). Following these initial reports, an epidemic swept through Europe affecting the racing, show and feral pigeon populations. The disease signs were consistent, and generally included a series of nervous disorders: bilateral or unilateral locomotor disturbances of wings and or legs, torticollis and watery green diarrhoea. If infected during breeding or moulting, increased embryo mortality or deformed feathers could be noted, respectively (Alexander et al., Citation1984c).
The mixing of pigeons in association with races and the extensive trade in these birds and their products is likely to be the cause of the rapid dissemination of this disease in racing pigeon communities. Despite a ban on the racing of pigeons from the Continent back to Great Britain, the disease reached England in June 1983, and by December 1983 it had been recorded in 29 counties across Great Britain (Alexander et al., Citation1984b).
From the outset, unpublished reports indicated that the virus isolated from the diseased pigeons was atypical, and distinguishable from the ‘classical’ avian paramyxovirus-1 (APMV-1). This variant nature of the APMV-1 causing disease in pigeons was confirmed by Alexander et al. (Citation1984b), where the authors report that the viruses isolated from racing pigeons (PPMV-1) in the United Kingdom were indistinguishable from those affecting pigeons on the continent; also that the monoclonal antibody binding pattern for these isolates was distinct from those for existing strains of APMV-1. Rapid typing and identification of PPMV-1 viruses were greatly enhanced by the development of a mouse monoclonal antibody (161/617) that, among APMV-1 viruses, bound exclusively to the PPMV-1 isolates (Collins et al., Citation1989).
The risk posed by the virus causing disease in pigeons to poultry became reality when in 1984 a virus serologically indistinguishable from the variant APMV-1 strain affecting pigeons was shown to be responsible for 22 outbreaks of Newcastle disease (ND) in poultry in the United Kingdom. It has been widely accepted that the virus was passed to poultry via feed contaminated with faeces from infected feral pigeons (Anonymous, Citation1984; Alexander et al., Citation1984a).
The true panzootic nature of this disease is demonstrated by the isolations reported from many countries world-wide (Alexander et al., Citation1985a; Shirai et al., Citation1986; Gelb et al., Citation1987; Ide, Citation1987; Pearson et al., Citation1987; Abu-Elzein et al., Citation1999; Zanetti et al., Citation2001). The origins of the panzootic in pigeons appeared to be in the Middle East during the late 1970s and evidence for this was the fortuitous isolation of a PPMV-1 virus in Iraq in 1978 (Kaleta et al., Citation1985). The panzootic peak was during the early 1980s and, although numbers of reported cases are now much lower now, it is still ongoing in Europe and other parts of the world.
The genetic and antigenic relationships between many different isolates of ND virus (not just PPMV-1) have been discussed (Collins et al., Citation1994;Ballagi Pordany et al., Citation1996; Alexander et al., Citation1997b; Lomniczi et al., Citation1998; Aldous et al., Citation2003). In all these studies, despite the PPMV-1 variant isolates locating together in their own discrete sublineage (VIb/4b) or antigenic group (P), they are reported to be closely related to other APMV-1 viruses. More specifically, the relationships between PPMV-1 isolates have also been investigated in a number of studies (Collins et al., Citation1996; Zanetti et al., Citation2001, Citation2003; Meulemans et al., Citation2002; Terregino et al., Citation2003; Ujvari et al., Citation2003).
In the present study, the phylogenetic relationships between 208 isolates from this ongoing panzootic of PPMV-1 were investigated based on the nucleotide sequence of a 375-nucleotide fragment at the 3′ end of the fusion protein (F) gene. This segment of the gene includes the region encoding the signal sequence and the precursor fusion protein cleavage activation site. As one of the major antigenic determinants of ND virus, the F gene is likely to display greater genetic variation than the internal genes. This characteristic is important for studying quite closely related populations, where a more conserved gene may show insufficient sequence variation to allow evolutionary hypotheses to be deduced.
Materials and Methods
Isolates
Virus isolates were grown in the allantoic cavities of 9-day-old to 10-day-old embryonated fowls’ eggs originating from a commercial specific pathogen free flock and were supplied by the International Reference Laboratory at VLA-Weybridge. The isolate details, their abbreviations and accession numbers are presented in . These viruses represent APMV-1 isolates from pigeons and show the monoclonal antibody (mAb) binding pattern associated with the panzootic viruses (Alexander et al., Citation1997b) or are viruses with that mAb pattern isolated from other bird species.
Table 1. Details of ND (APMV-1) viruses analysed in this study
Reverse transcription-polymerase chain reaction and polymerase chain reaction
Viral RNA was extracted from infective allantoic fluid using the Qiagen QIAamp viral RNA kit. The preparation of cDNA was carried out as described by Collins et al. (Citation1993), using an alternative cDNA primer (#MSF1), 5′-GACCGCTGACCACGAGGTTA. ‘Reddy-mix’ polymerase chain reaction (PCR) master mix (AB gene) was used for the PCR, using the cDNA forward primer and reverse primer (#2) 5′-AGTCGGAGGATGTTGGCAGC to generate an amplicon of approximately 700 base pairs. The final concentrations of components in the PCR mix were 1.25 u Taq DNA polymerase, 75 mM Tris–HCl, 20 mM (NH4)2SO4, 1.5 mM MgCl2, 0.01% (v/v) Tween 20, 1 μM each primer and 0.2 mM dATP, dCTP, dGTP and dTTP. The cycling parameters were: initial denaturation step of 1 min at 95°C; 30 cycles of 1 min at 94°C, 1 min at 50°C, 3 min at 72°C; and final extension step 72°C for 10 min. The PCR products were separated by gel electrophoresis using 2% w/v agarose gel in Tris-acetate buffer, stained with ethidium bromide and visualized under ultraviolet light. The product was excised and purified using QiaQuick (Qiagen) gel extraction kit.
Nucleotide sequencing
The forward primer used was (#7) 5′-TTAGAAAAAACACGGGTAGAA (Collins et al., Citation1996), and the reverse primer was the same as for the PCR. Nucleotide sequencing was carried out using ‘BigDye’ DNA sequencing kits and a PE Applied Biosystems 310 genetic analyser.
Sequence analysis
Nucleotide sequence analysis and alignment was carried out using Lasergene DNASTAR software version 5. The sequences were aligned to begin at the start codon (ATG at position 47) of the fusion protein gene and finished at base 422, which is just downstream from the cleavage activation site. The sequences were aligned using the Clustal V method in Megalign. Subsequent phylogenetic analysis was performed using PHYLIP phylogenetic inference package, version 3.57. The results are presented as unrooted maximum likelihood phylogenetic trees in which the branch lengths are proportional to the predicted number of substitutions.
Results
The radial phylogram in was generated from a data-set based on the partial fusion gene nucleotide sequences of 225 APMV-1 isolates. Included in this study are 47 published sequences, of which 17 represent other previously determined lineages of APMV-1 (Ballagi et al., Citation1996; Aldous et al., Citation2003) and 30 PPMV-1 isolates. The remaining 178 isolates were sequenced for this study and are all PPMV-1 isolates; determined by their binding pattern with a panel of monoclonal antibodies (). This initial tree () is provided as a visual guide to illustrate the overall relationship between subgroup 4b (PPMV-1 variants) isolates and the APMV-1 group as a whole. It is not provided to allow inferences to be made between individual isolates.
Figure 1. Unrooted maximum likelihood radial phylogram based on nucleotide sequence data from 225 APMV-1 isolates, including 208 PPMV-1 isolates and 17 representative of the other genetic lineages (Aldous et al., Citation2003). The region analysed was a 375 base pair fragment (47 to 422) at the 3′ end of the fusion protein gene. Branch lengths represent the predicted number of substitutions and are proportional to the differences between the isolates. The individual names of each PPMV-1 isolate included in this phylogram are not included. The groups selected for this study are shaded on the tree and labelled. The branch to isolate HFRDK77188 (group 6) is not drawn to scale; its actual branch length value is 1.66.
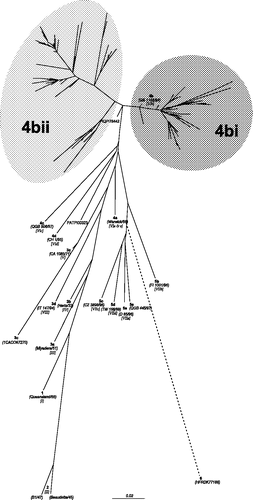
With the exception of one (PATPI00323), all isolates were grouped phylogenetically into lineage 4b (Aldous et al., Citation2003) synonymous with VIb (Lomniczi et al., Citation1998), which could be separated into two further groups (4bi and 4bii) (). These two groups have been analysed as individual data-sets and the trees generated from them are displayed in and respectively. In these two trees, identical sequences have not been labelled; in these cases a single representative sequence has been included and marked with an asterisk. The isolates with sequences identical to these asterisked isolates are presented in . For the ease of discussion only, the two subgroups, 4bi and 4bii have each been subdivided into three further clades, a, b, c and d, e, f, respectively. It is not proposed that these are to be considered taxonomical groupings.
Figure 2. Unrooted maximum likelihood radial phylogram of subgroup 4bi isolates, based on nucleotide sequence data from 91 PPMV-1 isolates. The region analysed was a 375 base pair fragment (47 to 422) at the 3′ end of the fusion protein gene. Branch lengths represent the predicted number of substitutions and are proportional to the differences between the isolates. * Isolates represent at least one other identical sequence as presented in .
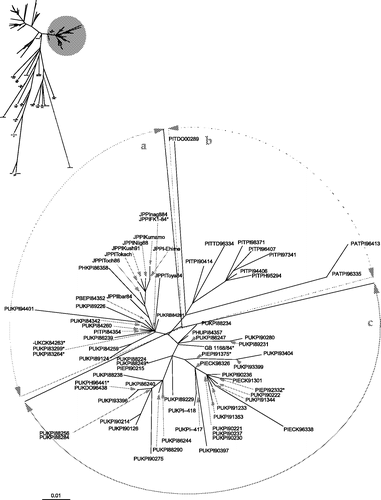
Figure 3. Unrooted maximum likelihood radial phylogram of subgroup 4bii isolates, based on nucleotide sequence data from 114 PPMV-1 isolates. The region analysed was a 375 base pair fragment (47 to 422) at the 3′ end of the fusion protein gene. Branch lengths represent the predicted number of substitutions and are proportional to the differences between the isolates. * Isolates represent at least one other identical sequence as presented in .
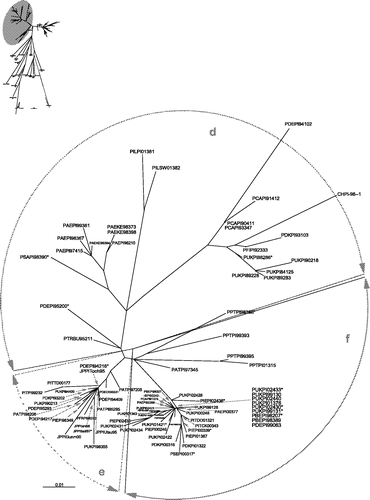
Table 2. Isolates included in and (marked *) that represent other isolates with identical sequences
Subgroup 4bi
The 4bi group is based on the sequences of 91 isolates (). Broadly, it is composed of isolates of Asian and European (mainly UK) origins obtained between 1983 and 2002. The majority of isolates were obtained from pigeon hosts, with a few from chickens (seven), doves (two), pheasants (two), a turtle dove, a fantail pigeon and from rice bran taken from a storage barn at Liverpool docks. Group 4bi can be divided into three clades a, b and c.
4bi a
All of the Asian (Japanese) isolates in this group fall into a discrete branch; other non-Asian isolates also stem from the same primary node. The rest are mainly UK strains isolated during the mid-1980s, although isolates from Belgium and Italy are also included.
4bi b
This group consists of a fairly diverse group of Italian (eight) and Austrian (two) isolates. These were all isolated between 1990 and 2000. The two Austrian isolates are on their own branch, as is a single Italian isolate that was obtained from a dove. The single virus in group 4bi b isolated from a pheasant in Italy in 1995 was closest to an Italian pigeon isolate obtained in 1994.
4bi c
A fairly diverse cluster of UK and Irish (and one Hungarian) isolates can be identified at the base of the 4bi c clade. All these isolates were obtained between 1984 and 1998. Included in this cluster are isolates from a pheasant, dove and fantail pigeon all obtained in 1996 from the same vicinity. These isolates were reported have an unusual mAb binding pattern (Alexander et al., Citation1997a). With the exception of three samples already mentioned and three isolations from Irish chickens, all isolates were obtained from pigeons.
Group 4bii
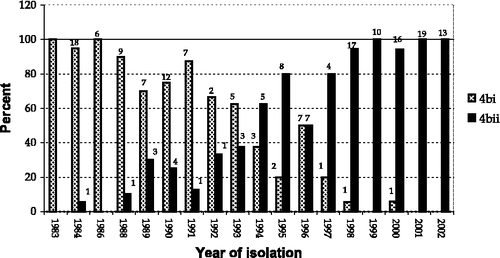
The group 4bii phylogram is based on the sequence for 114 isolates and can be further divided into three clades, d, e and f (). Broadly, this group includes isolates from Europe, North America and Asia (China and Middle East) obtained between 1984 and 2002. The trend of the distribution of isolates according to their year of isolation can be seen in . Clearly, group 4bii is the predominant strain in the latter period of this ongoing panzootic, with the number of type 4bi isolations diminishing from the late 1980s onwards. Again, the majority of isolations were made from pigeon hosts; but also from kestrels (Falco tinnunculus) (three), chickens (Gallus domesticus) (two), cockatoos (Cacatua spp) (two) and from a single budgerigar (Melopsittacus undulatus), turtle dove (Streptopelia risoria), dove, pheasant (Colchicus spp), swan (Cygnus spp), and falcon (Falco spp.).
Figure 4. Percentage distribution of isolates in group 4bi and group 4bii according to year of isolation (data label is actual number of isolations).
4bii d
Ten isolates with Middle Eastern origins cluster together exclusively on two branches in this clade; the two isolates from Israel are on a separate branch from those from the United Arab Emirates. Of these 10 Middle Eastern strains, seven were from pigeons, three from kestrels and one from a swan; all were isolated during the period from 1983 to 2001. The remaining isolates in this clade have less obvious links between their origins, being composed of isolates spanning three continents from two decades. A further node included in this group separates two isolates from 1995; one isolated from budgerigar in Turkey, the other from a pigeon in Germany.
4bii e
This clade of 21 mainly closely related viruses includes isolates from Asia and Europe obtained between 1990 and 2000; all except one from a turtledove were isolated from pigeons.
4bii f
The adjacent dense cluster included in clade 4bii f is comprised of European and Middle Eastern viruses isolated between 1998 and 2002 (with one exception from Canada putatively isolated in 1990). All the isolates in this cluster are from pigeons. Four Portuguese isolates form a further discrete branch within this group, all from pigeons and isolated between 1997 and 2001. Arising from the same node as these Portuguese isolates is an Austrian isolate obtained in 1997.
The trend of the distribution of isolates according to their year of isolation can be seen in . Clearly, group 4bi was the predominant strain at the beginning of the panzootic, but as the type 4bii viruses began to emerge in the late 1980s the group 4bi viruses began to decline.
Discussion
PPMV-1 isolates form a discrete group within the AMPV-1 population based on their genetic (lineage 4b/Vib) and antigenic (mAb P) properties. It was anticipated that since all isolates included in this study had been confirmed as mAb type P isolates (PPMV-1), all would fall into genetic lineage 4b.
Every effort has been made to include isolates in this study that are truly representative of the course of the whole panzootic. However, it is probable that some distortion of the results will have occurred due to availability of submissions from different countries and also the extent of disease surveillance being carried out. In spite of these caveats, there are a number of trends that can be identified and inferences made from the data presented here.
From the data obtained in this study it can be deduced that within this antigenically homogeneous group of viruses there is sufficient intra-group genetic variation to allow these isolates to be separated into two further groups within the 4b lineage of AMPV-1. The two groups (4bi and 4bii) proposed in this work were selected according to the topology of the initial phylogenetic tree (). The isolates falling within these two groups were analysed as separate data-sets of more practicable sizes, and in each of these the isolates tended to fall into three groups.
Of the 178 isolates analysed in this study only one (PATPI00323) was not placed in lineage 4b. When analysed phylogenetically with data from a previous study (Aldous et al., Citation2003) isolate PATPI00323 was found to fall into lineage 4a, most closely to another pigeon isolate (PTRPI95107) and two mAb group J isolates (JAESA90099 and JAEFA96038). Further analysis will be carried out on this isolate to determine whether it is a mixed infection, or whether it is a new virus with unusual antigenic properties. Another isolate, PIQPI78442, despite being located in lineage 4b, could not be allocated to either group 4bi or 4bii. The location of isolate PIQPI78442 in the phylogram at the base of the node from which the two groups 4bi and 4bii divide indicates the potential role of this, or a very similar, isolate as a possible progenitor virus of the PPMV-1 strain, which is in good agreement with previous studies where it is proposed as the first isolation of PPMV-1 (Kaleta et al., Citation1985).
There are two examples where monoclonal antibody binding patterns were used to investigate the source of outbreaks of ND in domestic poultry. In the first, virus isolated from contaminated food stores at Liverpool docks and from pigeons infesting them was confirmed to be antigenically similar to the virus responsible for outbreaks of ND in poultry in the United Kingdom at the same time, and was therefore proposed as the source (Alexander et al., Citation1985b). The results of the present study confirm this since the rice bran sample (PUKRB84261) and contemporaneous pigeon isolates are closely related to viruses isolated from chickens ( and a). In the second example from 1996, diseased pheasants were found to be infected with a PPMV-1 strain. When a virus with similar properties was isolated from pigeons and doves in the same locality during the same time period, these were proposed as the source for this outbreak (Alexander et al., Citation1997a). Verification of this is provided in the present study where the virus from the pheasant (PUKPH96441) is genetically identical to the pigeon and dove isolates, PUKPI96437 and PUKDO96438, obtained at the same time.
More than 43% of the isolates analysed in this study originated from the United Kingdom. Most of the isolates (>64%) in 4bi are from United Kingdom and Ireland. While UK and Irish isolates are present in 4bii they do not represent the majority (<32%). The bulk of the viruses in 4bi were isolated between the early 1980s and the early 1990s. Those in 4bii were generally isolated from the early to mid-1990s until 2002. The reasons for this progressive variation are not clear. The section of the genome used in this study is located in the fusion gene. This important surface glycoprotein is likely to show genetic variation due to selective or adaptive processes, and may well be affected by the use of vaccination. In Great Britain there was a non-vaccination policy at the time that the PPMV-1 strain emerged, but voluntary vaccination was allowed for racing pigeons from 1984, although it was not made compulsory until 1994 and only then for birds taking part in races. It is possible that 4bii strains emerged as a result of immunological selection processes induced by vaccination.
Despite the tendency of 4bi viruses to have been isolated at the beginning of the panzootic and 4bii viruses to have been isolated more recently (), there was no evidence of sequential progression amongst the six subgroups identified. There was evidence that viruses from several of the six subgroups could be circulating in the pigeon and dove populations of a country at the same time. For example, isolates originating from Italy in 2000 were from three separate subgroups (4bi b, 4bii e and 4bii f). This is in agreement with previous studies where it has been concluded that at any one time there are a number of genetically distinct ND virus pools circulating in a host population (Alexander, Citation2000).
The relatedness between isolates is influenced by their geographical origins, the full effect of which is likely to be masked by the actions of humans in the racing, showing, trade and transport of these birds and their products. Despite this, isolates originating from Portugal, United Arab Emirates and Italy are good examples of the geographical restriction to which these viruses appear to be subjected, where all occupy at least one specific discrete clade. In the case of Italy and United Arab Emirates, both countries are represented in other subgroups by isolates obtained during the same period. Isolates from the United Arab Emirates obtained during the late 1990s are only found in group 4bii. However, within this group there are two distinct strains present. One of which appears to be specific to the Middle East (4bii d), and the other more similar to a strain circulating in Northern Europe during the same period (4bii f). These two strains are likely to represent two separate introductions of the virus to the region, rather than concurrent evolution of two emerging strains. Another geographically specific example is that of the Portuguese isolates, which are only found on a separate branch in 4bii. In this case, there is an Austrian sample that is included in the cluster and predates the Portuguese isolates by one year. Conversely, geographical similarity does not hold true for the one Chinese isolate, since it is more similar to a Danish isolate than the other Asian isolates.
In conclusion, a single serotype of virus has been responsible for the panzootic in pigeons over the past two decades and has also caused disease in domestic poultry. During the course of the panzootic at least two genetically distinct strains of the virus have been identified that are separated chronologically; the viruses isolated during the first decade are predominately from group 4bi, while for the second decade type 4bii became the dominant strain. In addition to this chronological trend, geographical origins and host restrictions have also influenced the distribution of these viruses. The isolates in group 4bi are from eight different countries (with the majority being the United Kingdom and Ireland) and have been isolated from four different hosts; the isolates from 4bii are from many different countries (19) and eight different hosts.
The panzootic in pigeons and doves caused by PPMV-1 viruses is continuing in many countries of the world and represents an ongoing serious threat to domestic poultry, pigeons and other bird populations world-wide.
Acknowledgments
This work was supported financially by a Department of Environment, Food and Rural Affairs research grant. The authors thank Ruth Manvell for her advice and for supplying the virus isolates and their mAb binding details, and Ian Brown, Jill Banks and Mike Collins for their advice and help with preparation of the manuscript.
References
- Abu-Elzein , EME , Manvell , R , Alexander , DJ and Alafaleq , AI . 1999 . Pigeon paramyxovirus-1 (P group) as the cause of severe outbreaks in Fancy Columba livia in Saudi Arabia . Journal Veterinary Medicine , 46 : 689 – 692 .
- Aldous , EW , Mynn , JK , Banks , J and Alexander , DJ . 2003 . A molecular epidemiological study of avian paramyxovirus type 1 (Newcastle disease virus) isolates by phylogenetic analysis of a partial nucleotide sequence of the fusion protein gene . Avian Pathology , 32 : 239 – 258 .
- Alexander , DJ . 2000 . Newcastle disease and other avian paramyxoviruses . Revue Scientifique et Technique de L Office International des Epizooties , 19 : 443 – 462 .
- Alexander , DJ , Parsons , G and Marshall , R . 1984a . Infection of fowls with Newcastle disease virus by food contaminated with pigeon faeces . Veterinary Record , 115 : 601 – 602 .
- Alexander , DJ , Russell , PH and Collins , MS . 1984b . Paramyxovirus type 1 infections of racing pigeons: 1 characterisation of isolated viruses . Veterinary Record , 114 : 444 – 446 .
- Alexander , DJ , Wilson , GW , Thain , JA and Lister , SA . 1984c . Avian Paramyxovirus type 1 infection of racing pigeons: 3. Epizootiological considerations . Veterinary Record , 115 : 213 – 216 .
- Alexander , DJ , Russell , PH , Parsons , G , Abu-Elzein , E , Ballouh , A , Cernik , K , Engstrom , B , Fevereiro , M , Fleury , H , Guittet , M , Kaleta , E , Kihm , U , Kosters , J , Lomniczi , B , Meister , J , Meulemans , G , Nerome , K , Petek , M , Pokomunski , S , Polten , B , Prip , M , Richter , R , Saghy , E , Samberg , Y and Spanoghe , LTB . 1985a . Antigenic and biological characterisation of avian paramyxovirus type 1 isolates from pigeons—an international collaborative study . Avian Pathology , 14 : 365 – 376 .
- Alexander , DJ , Wilson , GW , Russell , PH , Lister , SA and Parsons , G . 1985b . Newcastle disease outbreaks in fowl in Great Britain during 1984 . Veterinary Record , 117 : 429 – 434 .
- Alexander , DJ , Manvell , RJ , Frost , KM , Pollitt , WJ , Welchman , D and Perry , K . 1997a . Newcastle disease outbreak in pheasants in Great Britain in May 1996 . Veterinary Record , 140 : 20 – 22 .
- Alexander , DJ , Manvell , RJ , Lowings , JP , Frost , KM , Collins , MS , Russell , PH and Smith , JE . 1997b . Antigenic diversity and similarities detected in avian paramyxovirus type 1 (Newcastle disease virus) isolates using monoclonal antibodies . Avian Pathology , 26 : 399 – 418 .
- Anonymous . 1984 . Newcastle disease: outbreaks linked to pigeons . Veterinary Record , 114 : 305 – 306 .
- Ballagi Pordany , A , Wehmann , E , Herczeg , J , Belak , S and Lomniczi , B . 1996 . Identification and grouping of Newcastle disease virus strains by restriction site analysis of a region from the F gene . Archives of Virology , 141 : 243 – 261 .
- Biancifiori , F and Fioroni , A . 1983 . An occurrence of Newcastle disease in pigeons: virological and serological studies on the isolates . Comparative Immunology and Microbiology of Infectious Diseases , 6 : 247 – 252 .
- Collins , MS , Alexander , DJ , Brockman , S , Kemp , PA and Manvell , RJ . 1989 . Evaluation of mouse monoclonal antibodies raised against an isolate of the variant paramyxovirus type 1 responsible for the current panzootic in pigeons . Archives of Virology , 104 : 53 – 61 .
- Collins , MS , Bashiruddin , JB and Alexander , DJ . 1993 . Deduced amino acid sequences at the fusion protein cleavage site of Newcastle disease viruses showing variation in antigenicity and pathogenicity . Archives of Virology , 128 : 363 – 370 .
- Collins , MC , Strong , I and Alexander , DJ . 1994 . Evaluation of the molecular basis of pathogenicity of the variant Newcastle disease viruses termed ‘pigeon PMV-1 viruses’ . Archives of Virology , 134 : 403 – 411 .
- Collins , MS , Strong , I and Alexander , DJ . 1996 . Pathogenicity and phylogenetic evaluation of the variant Newcastle disease virus termed ‘pigeon PMV-1 viruses’ based on the nucleotide sequence of the fusion protein gene . Archives of Virology , 141 : 635 – 647 .
- Gelb , J , Fries , PA and Peterson , FS . 1987 . Pathogenicity and cross protection of pigeon paramyxovirus 1 and Newcastle disease virus in young chickens . Avian Diseases , 31 : 601 – 606 .
- Ide , PR . 1987 . Virological studies of paramyxovirus type 1 infections in pigeons . Canadian Veterinary Journal , 28 : 601 – 603 .
- Kaleta , EF , Alexander , DJ and Russell , PH . 1985 . The first isolation of the avian PMV-1 virus responsible for the current panzootic in pigeons? . Avian Pathology , 14 : 553 – 557 .
- Lomniczi , B , Wehmann , E , Herczeg , J , Ballagi Pordany , A , Kaleta , EF , Werner , O , Meulemans , G , Jorgensen , PH , Mante , AP , Gielkens , AL , Capua , I and Damoser , J . 1998 . Newcastle disease outbreaks in recent years in Western Europe were caused by an old (VI) and a novel genotype (VII) . Archives of Virology , 143 : 49 – 64 .
- Meulemans , G , vandenBerg , TP , Decaesstecker , M and Boschmans , M . 2002 . Evolution of pigeon Newcastle disease virus strains . Avian Pathology , 31 : 515 – 519 .
- Pearson , JE , Senne , DA , Alexander , DJ , Taylor , WD , Peterson , LA and Russell , PH . 1987 . Characterization of Newcastle disease virus (avian paramyxovirus 1) isolated from pigeons . Avian Diseases , 31 : 105 – 111 .
- Shirai , J , Tsukamoto , K and Hihara , H . 1986 . Newcastle disease viruses isolated from racing pigeons in Japan . Nippon Juigaku Zasshi , 48 : 1091 – 1095 .
- Terregino , C , Cattoli , G , Grossele , B , Bertoli , E , Bertoli , E , Tisato , E and Capua , I . 2003 . Characterisation of Newcastle disease virus isolates obtained from Eurasian collard doves (Streptopelia decaocto) in Italy . Avian Pathology , 32 : 63 – 98 .
- Ujvari , D , Wehmann , E , Kaleta , EF , Werner , O , Savic , V , Nagy , E , Czifra , G and Lomniczi , B . 2003 . Phylogenetic analysis reveals extensive evolution of avian paramyxovirus type 1 strains of pigeons (Columba livia) and suggests multiple species transmission . Virus Research , 96 : 63 – 73 .
- Zanetti , F , Mattiello , R , Garbino , C , Kaloghlian , A , Terrera , MV , Boviez , J , Palma , E , Carrillo , E and Berinstein , A . 2001 . Biological and molecular characterization of a pigeon paramyxovirus type-1 isolate found in Argentina . Avian Diseases , 45 : 567 – 571 .
- Zanetti , F , Rodriguez , M , King , DJ , Capua , I , Carrillo , E , Seal , BS and Berinstein , A . 2003 . Matrix protein gene sequence analysis of avian paramyxovirus 1 isolates obtained from pigeons . Virus Genes , 26 : 199 – 206 .