Abstract
It has now been established that circovirus infection is common in farmed geese, but little is known about the clinicopathological significance of such infections. Ten clinically diseased geese suspected of being infected by circovirus were studied by in situ hybridization using a goose circovirus DNA probe. Circovirus DNA was demonstrated in the bursa of Fabricius (BF), spleen, thymus, bone marrow, liver, kidney, lung and heart, indicating that infection can be multisystemic. In some birds, virus DNA was present in very large quantities, most notably in the BF, liver and small intestine. With the exception of BF and thymus, there were no histological findings that would have suggested the presence of such quantities of circovirus DNA. In view of the very large quantities of virus DNA labelling present in some tissues, and by analogy to porcine circovirus type 2 infection and psittacine beak and feather virus infections, which are known to cause severe disease, and which have similar virus distribution to that found in our geese, it seems probable that the circovirus was important in the disease manifestations shown by the infected geese.
Etude de circovirus, par hybridation in situ, chez des oies infectées
Il est maintenant établi que l'infection à circovirus est commune dans les élevages d'oie, mais on sait peu de chose sur l'importance clinicopathologique de telles infections. Dix oies cliniquement malades, suspectées d’être infectées par le circovirus, ont fait l'objet d'une étude par hybridation in situ en utilisant une sonde ADN circovirus de l'oie. L'ADN du circovirus a été mis en évidence dans la bourse de Fabricius (BF), la rate, le thymus, la moelle osseuse, le foie, les reins, les poumons et le cœur, indiquant que l'infection peut être multisystémique. Chez quelques animaux, l'ADN viral a été mis en évidence en très grande quantité, plus particulièrement dans la BF, le foie, et l'intestin grêle. A l'exception de la BF et du thymus, les résultats histologiques n'ont pas permis de suggérer la présence de telle quantité d'ADN de circovirus. Etant donné la très grande quantité d'ADN présent dans quelques tissus, et par analogie à l'infection due au circovirus porcin de type 2 ainsi qu'aux infections virales du bec et des plumes des psittacidés qui sont connues pour entraîner des maladies graves, et qui ont une distribution virale similaire à celle observée chez les oies, il semble probable que le circovirus ait été important dans les manifestations de la maladie exprimée chez les oies infectées.
Untersuchung einer Circovirusinfektion in Gänsen mittels in situ-Hybrisidation
Es ist mittlerweile nachgewiesen, dass die Circovirusinfektion in Gänseherden weit verbreitet ist. Wenig ist jedoch über die klinisch-pathologische Bedeutung einer solchen Infektion bekannt. Zehn klinisch kranke Gänse, bei denen eine Circovirusinfektion vermutet wurde, wurden mittels in situ-Hybridisation unter Verwendung einer Circovirus-DNS-Probe untersucht. Circovirus-DNS wurde in Bursa Fabricii (BF), Milz, Thymus, Knochenmark, Leber, Niere, Lunge und Herz nachgewiesen, was darauf hin weist, dass die Infektion multisystemisch sein kann. In einigen Tieren war der virale DNS-Gehalt sehr groß, insbesondere in BF, Leber, Dünndarm. Mit Ausnahme von BF und Thymus gab es keine histologischen Veränderungen in den Organen, die das Vorkommen so großer Circovirus-DNS-Mengen hätten vermuten lassen. Angesichts der sehr großen Mengen markierter Virus-DNS in einigen Organen und in Analogie zu Infektionen mit Circovirus Typ 2 beim Schwein und mit psittazidem Schnabel- und Federnvirus, die als Erreger schwerer Erkrankungen bekannt sind und eine der hier bei den Gänsen gefundene ähnliche Virusverteilung im Körper haben, scheint es wahrscheinlich, dass das Circovirus eine bedeutsame Rolle bei dem von den infizierten Gänsen gezeigten Krankheitsbild spielte.
Estudio de gansos infectados con circovirus mediante hibridación in situ
Aunque ya se ha determinado que la infección por circovirus es común en gansos de granja, se desconoce en gran medida la significación clinicopatológica de tales infecciones. Diez gansos clínicamente enfermos y sospechosos de estar infectados con circovius fueron estudiados mediante hibridación in situ utilizando una sonda de ADN frente a circovirus de ganso. El ADN de circovirus se detectó en bolsa de Fabricio (BF), bazo, timo, médula ósea, hígado, riñón, pulmón y corazón, lo cual indica que la infección era multisistémica. En algunas aves, el ADN vírico se encontraba en grandes cantidades, mayoritariamente en BF, hígado e intestino delgado. Con la excepción de la BF y el timo, no se observaron hallazgos microscópicos que sugirieran la presencia de tales cantidades de ADN de circovirus. A la vista de las grandes cantidades de ADN vírico presente en algunos tejidos, y por analogía con las infecciones por circovirus porcino tipo 2 y el virus de la enfermedad del pico y las plumas que se conoce que causan enfermedades graves, y que presentan una distribución del virus similar a la encontrada en estos gansos, parece probable que el circovirus tuviera un papel relevante en la manifestación de la enfermedad mostrada por los gansos infectados.
Introduction
Circoviruses are small, non-enveloped, spherical viruses (diameter 15 to 25 nm) that contain a circular single-stranded DNA genome. The genus Circovirus of the family Circoviridae contains, or is likely to contain, porcine circovirus type 1 and porcine circovirus type 2 (PCV2), psittacine beak and feather disease virus (BFDV) and pigeon or columbid circovirus (PiCV) (Mankertz et al., Citation2000; Todd, Citation2000; Todd et al., Citation2001b). Additional candidate members of the genus have also recently been described, namely canary circovirus (Todd et al., Citation2001a), goose circovirus (GoCV) (Todd et al., Citation2001b) and duck circovirus (Hattermann et al., Citation2003).
With respect to the pathogenesis and significance of circovirus infections, most is known about the porcine circoviruses and BFDV infection, and there is mounting evidence that the circoviruses cause immunosuppression and disease. In the case of geese infected by circovirus, relatively little information is available. The viruses were first found in inclusion bodies in the bursa of Fabricius (BF) of geese affected by a runting syndrome (Soike et al., Citation1999). There was associated lymphocyte depletion. Virus particles were also found in the spleen and thymus of some birds by negative contrast electron microscopy. Goose circoviruses have been recovered from geese in Taiwan suffering feathering abnormalities and stunting (Chen et al., Citation2003), but no other clinical or pathological information is available. In a recent study, Hungarian geese flocks diagnosed as infected by polymerase chain reaction (PCR) or dot blot hybridization were compared with non-infected flocks. With the exception of circovirus inclusions, which were found in the BF of birds in three infected flocks, no particular clinical syndrome or laboratory findings could be specifically associated with circovirus infection (Ball et al., Citation2004). Thus, there is a need for further studies to establish the role, if any, of circoviruses in disease in geese.
In situ hybridization (ISH) has proven a useful tool in the study of many infectious diseases. In the case of circovirus infections, for example, examination of infected pigeons by ISH revealed that circovirus DNA was present in many tissues, sometimes in large quantities, without its presence having been suspected previously (Smyth et al., Citation2001).
The present paper describes a study of the distribution of circovirus DNA in infected geese by ISH and its association with pathological changes.
Materials and Methods
Geese and examinations
Tissues were available from 10 geese for examination. One goose was 2 weeks old, and the others ranged from 8 to 10 weeks old. The 10-week-old geese had a history of enteritis and “apathy”. All of the other geese had a history of runting (). The gross and histological findings have already been described for the runted geese (Soike et al., Citation1999). Briefly, lymphocytic depletion and histiocytosis were seen in the BF and randomly dispersed inflammatory infiltrates were found in the lung, liver or myocardium of individual birds. Ultrastructural examination of inclusions in the BF revealed paracrystalline arrays of circovirus-like viruses. Negative contrast electron microscopy (EM) had revealed circovirus-like particles in the BF of all the eight birds in that study, in the spleen of four birds and in the thymus of two birds.
Table 1. Results of examination of tissues from diseased geese for circovirus by in situ hybridization: where available, results of negative contrast electron microscopic examination are given in parentheses
In the present study, formalin-fixed, paraffin-embedded tissues from 10 geese were studied by ISH. These included BF from seven birds, spleen from seven birds, thymus from five birds, liver from 10 birds, kidney from seven birds, small intestine from nine birds, bone marrow from four birds, lung from seven birds, heart from six birds and brain from seven birds.
DNA probes and ISH
The GoCV-specific probe was prepared from a plasmid vector containing cloned GoCV-specific DNA. Plasmid GoCV-IP (Todd et al., Citation2001b) contained a 1.8 kbp fragment equivalent to the complete GoCV genome that had been amplified from a clinical sample (1394/97) using inverse primers (IP). The probe used for ISH was produced by direct DIG-labelling of the 1.8 kbp GoCV-specific DNA, generated by PCR. PCR was performed using the GoCV-IP plasmid as a template and the inverse primers at 94°C for 30 sec, at 60°C for 30 sec, and at 68°C for 2 min for 35 cycles.
A “control” probe was prepared by labelling the pCR2.1 TOPO plasmid (with no circovirus DNA insert) using the random prime method (Smyth et al., Citation2001).
ISH was performed as described by Smyth et al. (Citation2001) using low stringency conditions. The intensity and extent of circovirus DNA labelling was graded on a scale from negative through to 5+. 1+ represents up to 10 infected cells per section, and 2+ represents 11 to 30 infected cells per tissue section. 3+ represented tissues where at least five infected cells were found in most x 20 fields or where several heavily infected foci were present in a tissue. 5+ represented tissues where greater than 20 infected cells were found in most x 20 fields.
Specificity and sensitivity testing for ISH
The goose circovirus probe was tested by ISH under conditions of low stringency on sections of tissues known to be infected with PCV2, BFDV, chicken anaemia virus, egg drop syndrome virus (EDSV), ovine adenovirus, bovine adenovirus type 4 (BAV-4) and bovine adenovirus type 10 (BAV-10), phocine distemper virus, bovine respiratory syncytial virus, paramyxovirus-1-infected BF, infectious bursal disease virus-infected BF, Aujeszky's disease virus-infected brain, chicken intestine infected with Eimeria acervulina or Eimeria maxima, and naturally occurring cases of coccidiosis in cattle, sheep, pigs, chickens and rabbits.
Also for control purposes, goose tissues giving strong positive hybridization signal with the goose circovirus probe were tested with two inappropriate probes (a probe to BAV-4 and an EDSV probe). As a further control, some tissues were subjected to ISH protocol except that either the control probe or no probe was added.
Comparison of ISH and negative contrast EM findings
A summary of negative contrast EM findings for these geese was previously reported (Soike et al., Citation1999). The results are presented in full in to enable comparison with ISH findings.
Results
Sensitivity and specificity of the ISH technique
Only tissues from the geese under study hybridized with the goose circovirus probe. The probe did not hybridize with any of the tissues infected with BFDV, PCV2, chicken anaemia virus, ovine adenovirus, BAV-10, BAV-4, EDSV, bovine respiratory syncytial virus, phocine distemper virus, Aujeszky's disease virus, and a range of gut tissues infected with a range of coccidial species.
Goose tissues that reacted positively with the goose circovirus probe did not react with BAV-4 or EDSV probes, or with the control plasmid probe.
ISH on test geese
Bursa of Fabricius
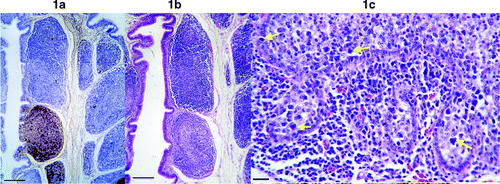
All seven BF hybridized with the goose probe. Very intense and extensive labelling of DNA was detected in the BF of five geese (). Infected cells were found both in the cortex and medulla, and circovirus DNA was detected both within nuclei and cytoplasm of infected cells. Some follicles were very heavily infected whereas other follicles contained either no or relatively few infected cells (). Much more virus DNA was present than had been suspected from the appearance of adjacent haematoxylin and eosin (H&E) sections. Cells containing intranuclear circovirus DNA appeared to be enlarged lymphocytes or lymphoblasts. Examination of adjacent H&E-stained sections revealed that moderate to heavily infected follicles were noticeably less densely populated with lymphocytes compared with uninfected or lightly infected follicles (). Large degenerate cells were sometimes observed in follicles () but these were usually few in number compared with the number of infected cells. Cells containing botryoid cytoplasmic inclusions were uncommon, even in heavily infected areas.
Figure 1. 1a: Low magnification picture of the BF showing circovirus DNA (labelled brown). Some follicles are heavily infected while others are uninfected. Infected cells are present in both the cortex and medulla of follicles. Bar=100 μm. 1b: H&E-stained section of BF adjacent to the section in 1a. The heavily infected follicle is less densely populated by lymphocytes, thus appearing less intensely stained. Bar=100 μm. 1c: High magnification of a heavily infected follicle showing large degenerate cells (arrows). Bar=12 μm.
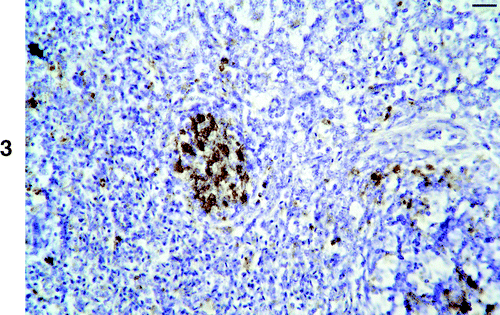
Thymus and spleen
Large numbers of positive cells were found in the thymus of two of the five birds examined (). Positive cells were present in both the cortex and in the medulla. Labelling was mainly intranuclear and in small to large lymphocytes (). There was also labelling in large cells thought to be swollen lymphoblasts and/or reticular cells (). Occasional very large structures whose nature could not be determined were also labelled (). There was marked depletion of the thymic cortex in one of these birds.
Figure 2. 2a: Low magnification of an infected thymus showing virus positive cells in both the cortex and medulla. Bar=100 μm. 2b: High magnification of the thymus showing nuclear labelling of lymphocytes (arrows), dense labelling in very enlarged cells (white arrowhead) and in an unidentified structure (see results) (black arrowhead). Bar=10 μm.
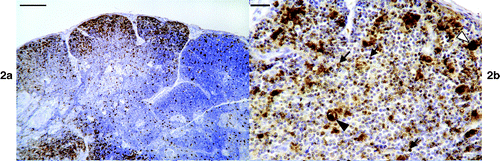
The spleen was positive in six of the seven birds examined. Randomly distributed cells were present throughout the spleen in all of these cases. One spleen was quite heavily infected, with virus DNA concentrated in the germinal centres (). No pathological change was recognized in these germinal centres in adjacent H&E sections.
Liver and kidney
Circovirus DNA was found in the liver of all 10 birds, although in three birds positive cells were very rare. Four birds contained numerous positive cells in the liver (). These cells were mainly within sinusoids and examination of adjacent H&E sections would suggest these may have been Kupfer cells. Labelling was mainly intranuclear but cytoplasmic labelling was also common. In the portal areas, cells resembling macrophages were positive. Apart from occasional vacuolated cells, some of which contained faint golden brown material, and occasional degenerate cells, no morphological abnormality was found in heavily infected areas.
Figure 4. ISH on liver showing abundant virus DNA, which is mainly present in sinusoidal lining cells. Bar=10 μm.
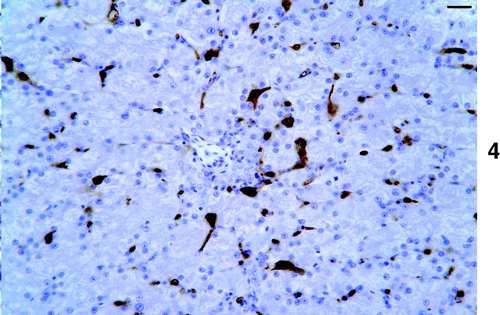
Positive cells were found in six of the eight kidneys examined, although positive cells were infrequent and mainly found randomly distributed in the renal interstitium.
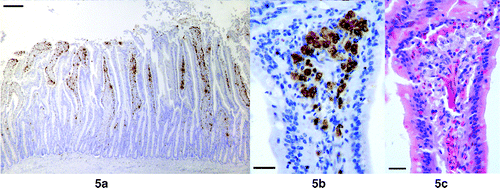
Small intestine
Positive cells were found in the small intestine of all eight birds examined, and were present in very large numbers in five birds. They were found mainly in the lamina propria of the villi and mainly subepithelially (,). However, positive cells were found within the epithelium and occasionally in intercrypt tissue. Labelling was mostly cytoplasmic but nuclear labelling was also observed. Examination of adjacent H&E sections revealed that many of the labelled cells in the lamina propria had very pale staining cytoplasm and open faced nuclei, and they resembled macrophages ().
Other tissues
Two of the four bone marrows examined contained positive cells. Positive cells were found either within lymphoid aggregates or as large scattered individual intrasinusoidal or extrasinusoidal cells.
Five of six heart samples contained positive cells. These were usually single cells within the interstitium with nuclear labelling, but occasionally several positive cells could be found within areas containing inflammatory cell infiltrates. Examination of adjacent H&E sections of the latter revealed non-suppurative myocarditis. Within affected areas there were several enlarged (and sometimes irregularly shaped) cardiomyocyte nuclei, but these were unlabelled by the circovirus probe.
Positive cells were found in six of the seven lung samples. These were usually found in areas of inflammatory cell infiltration.
Circovirus-infected cells were not detected in any of the seven brains examined
BF was not present from the two 10-week-old birds but circovirus DNA could be detected in most of the other tissues examined from these birds.
Comparison of ISH and negative contrast EM findings
For all tissues where both tests were applied, tissues found positive by EM were positive by ISH (). However, some tissues found positive by ISH were negative by EM ().
Discussion
A digoxygenin DNA probe was prepared from goose circovirus DNA and shown to specifically hybridize with circovirus DNA in infected geese. The probe did not cross-hybridize with other circoviruses such as BFDV, PiCV and PCV2 under low stringency conditions, suggesting that the probe only hybridizes with goose circovirus or very closely related circoviruses. This probe was applied to a range of tissues from diseased geese by ISH. Negative-contrast EM examination had revealed circovirus-like viruses in the BF of all birds, in the spleen of four birds and in the thymus of two birds (Soike et al., Citation1999; Soike, unpublished observations).
The ISH study, however, showed that goose circovirus DNA could be found in all tissue types tested with the exception of brain, and there was extensive labelling of virus DNA in the tissues of some of these birds. Virus DNA was most abundant in the BF, liver and small intestine. ISH was more sensitive than negative contrast EM. While botryoid intracytoplasmic inclusion bodies were detected in the BF of some of the birds, ISH was able to detect the presence of circovirus DNA in BF that did not contain the characteristic inclusions. Furthermore, ISH revealed much more virus DNA than had been expected on the basis of the histological appearance of the BF. Extensive labelling of virus DNA was also found in the liver and small intestine of some birds. Again, the presence of circovirus had not been suspected on the basis of the histological appearance. Careful retrospective examination of adjacent H&E sections of these livers and small intestines revealed either subtle histological changes or no recognizable change. Where present, changes were less pronounced than those found in PiCV-infected pigeons (Smyth et al., Citation2001). It is suggested that such changes where detected should raise suspicion of circovirus infection, but they could not be viewed as being pathognomonic.
The amount of circovirus DNA present in the liver and small intestine would suggest that these may be useful tissues to examine for the presence of circovirus, when BF is not available. However, in young birds the BF is a key tissue that should be examined before conclusions can be drawn about negativity, or low virus load. Examination of liver and small intestine may also be useful in older birds where the BF has undergone the normal age-associated regression. The amount of circovirus DNA in the small intestine may indicate that significant virus replication occurs in the gut, and it is possible that this may contribute to faecal, and thus environmental, contamination. It may also be of significance in terms of disease pathogenesis. The main clinical presentation in the study birds was runting, which could be a consequence of compromised gut function. It seems possible that the large amount of virus DNA and associated cellular distortion/infiltration could have been interfering with gut function in these birds. However, other mechanisms are also quite possible given the widespread distribution of infected cells within the body, and the very large numbers of infected cells in some tissues (e.g. liver). In this respect, it is interesting that PCV2 infection in pigs is associated with poor growth and wasting of affected pigs and, similar to the geese in the present study, PCV2 is widespread in the tissues of affected pigs (Rosell et al., Citation1999; Kennedy et al., Citation2000).
As was the case in pigeons, a very large amount of DNA was often present in the liver. It was also noteworthy that moderate to large amounts of DNA were detectable in the thymus, and that in one of these cases there was marked depletion of the thymic cortex.
The finding of virus DNA, often in large quantities, in a wide range of tissues has also been reported in birds infected with BFDV and PiCV, respectively, and in pigs infected with PCV2. These infections are now widely accepted as being highly significant, and therefore similar findings in geese raise the possibility that GoCV infection may well have clinicopathological significance. Since it has recently been shown that circovirus infection is widespread in farmed geese (Ball et al., Citation2004), there is clearly a need for further work to accurately establish the significance of this infection.
In conclusion, a sensitive and specific method of detecting goose circovirus DNA in formalin-fixed paraffin-embedded tissues has been developed. Using this technique, we have been able to demonstrate that circovirus can cause systemic infections in geese and that large quantities of virus DNA may be present. In this respect, goose circovirus is similar to the studied circoviruses associated with disease in other species (i.e. pigs, parrots and pigeons), and therefore may well be responsible for the disease present in our geese. The present study has also shown that circovirus DNA was present in tissues where it had not been suspected on the basis of histological examination. Careful comparison with adjacent H&E sections revealed only very subtle associated morphological changes in some tissues, and these could not be viewed as pathognomonic for circovirus infection. Thus, histological examination alone will not enable detection of the infection in many cases. Since infection of geese by circovirus is common, controlled studies are needed to fully establish the clinical significance of infection.
Translations of the abstract in French, German and Spanish are available on the Avian Pathology website.
References
- Ball , NW , Smyth , JA , Weston , JH , Borghmans , BJ , Palya , V , Glavitis , R , Ivanics , E , Dan , A and Todd , D . (2004) . Diagnosis of goose circovirus infection in Hungarian geese samples using polymerase chain reaction and dot blot hybridization tests . Avian Pathology , 33 : 51 – 58 .
- Chen , C-L , Chang , P-C , Lee , M-S , Shien , J-H , Ou , S-J and Shieh , HK . (2003) . Nucleotide sequences of goose circovirus isolated in Taiwan . Avian Pathology , 32 : 165 – 171 .
- Hattermann , K , Schmitt , C , Soike , D and Mankertz , A . (2003) . Cloning and sequencing of Duck circovirus (DuCV) . Archives of Virology , 148 : 2471 – 2480 .
- Kennedy , S , Moffett , D , McNeilly , F , Meehan , B , Ellis , J , Krakowka , S and Allan , GM . (2000) . Reproduction of lesions of postweaning multisystemic wasting syndrome by infection of conventional pigs with porcine circovirus alone or in combination with porcine parvovirus . Journal of Comparative Pathology , 122 : 9 – 24 .
- Mankertz , A , Hattermann , K , Ehlers , B and Soike , D . (2000) . Cloning and sequencing of columbid circovirus (CoCV), a new circovirus from pigeons . Archives of Virology , 145 : 1 – 11 .
- Rosell , C , Segales , J , Plana-Duran , J , Balasch , M , Rodriguez-Arrioja , GM , Kennedy , S , Allan , GM , McNeilly , F , Latimer , KS and Domingo , M . (1999) . Pathological, immunohistochemical and in-situ hybridization studies of natural cases of postweaning multisystemic wasting syndrome (PMWS) in pigs . Journal of Comparative Pathology , 120 : 59 – 78 .
- Smyth , JA , Weston , J , Moffett , DA and Todd , D . (2001) . Detection of circovirus infection in pigeons by in situ hybridization using cloned DNA probes . Journal of Veterinary Diagnostic Investigation , 13 : 475 – 482 .
- Soike , D , Köhler , B and Albrecht , K . (1999) . A circovirus-like infection in geese related to a runting syndrome . Avian Pathology , 28 : 199 – 202 .
- Todd , D . (2000) . Circoviruses: immunosuppressive threats to avian species: a review . Avian Pathology , 29 : 373 – 394 .
- Todd , D , Weston , J , Ball , NW , Borghmans , BJ , Smyth , JA , Gelmini , L and Lavazza , A . (2001a) . Nucleotide sequence-based identification of a novel circovirus of canaries . Avian Pathology , 30 : 321 – 325 .
- Todd , D , Weston , JH , Soike , D and Smyth , JA . (2001b) . Genome sequence determinations and analyses of novel circoviruses from goose and pigeon . Virology , 286 : 354 – 362 .