Abstract
A virus (AV71/99) was isolated from a green-cheeked Amazon parrot by propagation and passage in both primary embryo liver cells derived from blue and yellow macaw (Ara ararauna) embryos and chicken embryo liver cells. Electron microscopic examination of cytopathic agents derived from both types of cell cultures suggested that it was a coronavirus. This was confirmed using a pan-coronavirus reverse transcriptase polymerase chain reaction that amplified part of gene 1 that encodes the RNA-dependent RNA polymerase. The deduced sequence of 66 amino acids had 66 to 74% amino acid identity with the corresponding sequence of coronaviruses in groups 1, 2 and 3. Several other oligonucleotide primer pairs that give PCR products corresponding to genes 3, 5, N and the 3′-untranslated region of infectious bronchitis virus, turkey coronavirus and pheasant coronavirus (all in group 3) failed to do so with RNA from the parrot coronavirus. This is the first demonstration of a coronavirus in a psittacine species.
Isolement d'un coronavirus d'un perroquet amazone à joues vertes (Amazon viridigenalis Cassin)
Un virus (AV71/99) a été isolé d'un perroquet amazone à joues vertes sur des cultures primaires d'hépatocytes d'embryon d'ara bleu (Ara ararauna) et d'hépatocytes d'embryon de poulet, en réalisant des passages. L'examen en microscopie électronique des agents cytopathogènes dérivés des deux types de culture cellulaire a suggéré la présence d'un coronavirus. Ceci a été confirmé en utilisant les réactions de transcription inverse et d'amplification en chaîne par polymérase qui ont amplifié une partie du gène 1 qui code l'ARN polymérase ARN dépendante. La séquence déduite de 66 acides aminés présentait 66 à 74% d'identité en acides aminés avec la séquence correspondante des coronavirus des groupes 1, 2, et 3. Plusieurs autres paires d'amorce oligonucléotidique qui donnent des produits PCR correspondant aux gènes 3, 5, N et à la région 3' non traduite du virus de la bronchite infectieuse, du coronavirus de la dinde et du coronavirus du faisan (tous du groupe 3) n'ont pu le faire avec l'ARN du coronavirus du perroquet. C'est la première mise en évidence d'un coronavirus chez les psittacidés.
Isolierung eines Coronavirus aus einer Grünwangen-Amazone (Amazona viridigenalis Cassin)
Aus einer Grünwangen-Amazone (Amazon viridigenalis Cassin) wurde durch Anzüchtung und Passagierung in primären Embryoleberzellen von blauen und gelben Aras (Ara ararauna) sowie in Hühnerembryoleberzellen ein Virus isoliert (AV71/99). Die elektronenmikroskopische Untersuchung der zytopathogenen Agens aus beiden Zellkulturtypen ließ vermuten, dass es sich um ein Coronavirus handelte. Dieses wurde mittels einer für alle Coronaviren spezifischen Reverse Transkriptase- Polymerasekettenreaktion, die Teile des für die RNS-abhängige RNS-Polymerase kodierende Gen 1 amplifizierte. Die abgeleitete Sequenz aus 66 Aminosäuren wies eine 66–74%ige Aminosäurenübereinstimmung mit den entsprechenden Sequenzen der Coronaviren der Gruppen 1, 2 und 3 auf. Mehrere andere Oligonukleotidprimerpaare, die den Genen 3, 5, N und der nicht translatierten 3‘-Region des infektiösen Bronchitisvirus, des Putencoronavirus und des Fasanencoronavirus (alle in Gruppe 3) entsprechende PCR-Produkte amplifizieren, gelang dies nicht mit der RNS aus dem Papageiencoronavirus. Dies ist die Erstbeschreibung eines Coronavirus in einer Psittazidenspezies.
Aislamiento de un coronavirus de un loro Amapola (Amazon viridigenalis Cassin)
Se aisló un virus (AV71/99) de un loro Amapola mediante propagación y pases en células primarias de hígado de embrión derivadas de embriones de guacamayo azul y amarillo (Ara ararauna) y en células hepáticas de embrión de pollo. Los estudios en microscopio electrónico de los agentes citopáticos procedentes de ambos tipos de cultivos celulares sugirieron que se trataba de un coronavirus. Esto se confirmó mediante una transcriptasa reversa-reacción en cadena de la polimerasa de pan-coronavirus que amplificaba parte del gen 1 que codifica la RNA polimerasa-RNA dependiente. La secuencia deducida de 66 aminoácidos tenía entre el 66 al 74% de similitud aminoacídica con la correspondiente secuencia de los grupos 1, 2 y 3 de Coronavirus. Otros pares de cebadores oligonucleotídicos, que producen productos de PCR correspondientes a los genes 3, 5, N y a la región no codificantes 3’ del virus de la bronquitis infecciosa, del coronavirus del pavo y del coronavirus del faisán (todos ellos en el grupo 3), no amplificaron en el caso del coronavirus del loro. Esta es la primera descripción de un coronavirus en especies de psitácidas.
Introduction
During an investigation into the aetiological role of viruses in psittacine proventricular dilatation disease (PDD), formerly known as Macaw wasting disease, samples were received in December 1999 from a male green-cheeked Amazon parrot (Amazon viridigenalis Cassin). The bird had died in a poor condition following a history of anorexia, regurgitation and passing undigested food. At post mortem examination the main feature was a thin-walled grossly dilated proventriculus/ventriculus. Following histopathology examination of sections of crop and proventriculus, lesions similar to, but not conclusive of, PDD were found. In proven cases of PDD a non-suppurative encephalomyelitis and ganglioneuritis is seen in the brain and spinal cord (Gerlach, Citation1994). Unfortunately these organs were not available for examination. We subsequently attempted virus isolation from tissues that were available, which has led to the discovery and isolation of a coronavirus, the first one reported for a psittacine species.
Materials and Methods
Virus isolation
Samples of the proventriculus, heart, liver, spleen, pancreas, kidneys and intestinal contents were processed and inoculated onto cell cultures as described (Gough et al., Citation1988). Primary cell cultures were prepared from the livers of 15-day-old specific pathogen free (SPF) chicken embryos and 18-day-old blue and yellow macaw (Ara ararauna) embryos. The samples were also inoculated into embryonated SPF chicken eggs by both allantoic cavity and yolk-sac routes.
Electron microscopy
Inoculated cultures of liver cells from blue and yellow macaw and chicken embryos were frozen after 2 to 3 days and cell lysates prepared by two cycles of freezing and thawing. The cell debris was then removed following centrifugation at 720×g for 5 min and the clarified supernatant centrifuged at 32 000×g for 1 h at 4°C. The pellet was resuspended in a minimum volume of deionized water and examined by negative contrast electron microscopy. Uninoculated control cultures of macaw and chicken embryo liver cells received three blind passages and were similarly examined by electron microscopy.
Serology
As electron microscopy had revealed the presence of coronavirus-like particles in inoculated cell cultures, monospecific antiserum against the agent, designated AV71/99, was prepared in six SPF chickens. Briefly, each bird was inoculated at 10 days of age with 103.0 median tissue culture infectious doses (TCID50) of virus propagated in chicken embryo liver (CEL) cells, by the intranasal route. After 3 weeks the birds were re-inoculated with a similar dose of virus by the intravenous route. Blood samples were taken 2 weeks later and the separated sera tested for neutralizing antibodies to the coronavirus-like agent. The results indicated a very poor neutralizing antibody response so the birds were further inoculated by the intramuscular route with approximately 104.0 TCID50 of virus suspended in an oil emulsion. After a further 2 weeks the serum from each bird was tested and samples with similar neutralizing titres were pooled and used in haemagglutination inhibition (HI), virus neutralization (VN) and agar gel precipitation tests as described by Gough et al. (Citation1996). For the HI tests, antigens were prepared (OIE, Citation2004) from several infectious bronchitis virus (IBV) reference strains from chickens (M41, 793B, D274 and D1466), and two pheasant coronavirus isolates (ph/UK/438/94 and ph/UK/602/95) (Cavanagh et al., Citation2002), and used in one-way HI tests.
A total of 53 sera were examined for neutralizing antibodies to the coronavirus-like agent from the following psittacine birds; various species of macaws (42 sera), budgerigars (Melopsittacus undulatus) (three sera), sulphur-crested cockatoos (Cacatua galerita) (two sera), a Patagonian conure (Cyanoliseus patagonus) (one sera) and unspecified parrots (five sera). Briefly, doubling dilutions of serum were tested using 100 TCID50 virus derived from the fourth CEL passage of virus, in 96-well microtitre plates. The cultures were examined daily for cytopathic effect (CPE), and after 6/7 days the neutralization titres were calculated and expressed as the log2 reciprocal of the highest dilution of sera completely inhibiting CPE.
Reverse transcriptase-polymerase chain reactions
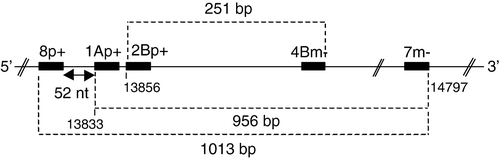
Viral RNA was extracted from a pool of clarified CEL cell lysate and culture supernatant using an RNeasy mini kit (Qiagen). Amplification of the RNA was carried out by reverse transcriptase-polymerase chain reactions (RT-PCRs) using Ready-to-go one-step RT-PCR beads (Amersham Biosciences) according to manufacturer's instructions. The pan-coronavirus, degenerate oligonucleotide primers 8p, 1Ap and 2Bp (genome sense) and 4Bm and 7m (antigenome sense) described by Stephensen et al. (Citation1999) were initially used. They correspond to sequences within the highly conserved (throughout the coronaviruses) RNA-dependent RNA polymerase (RdRp)-encoding region of gene 1b (). Subsequently several other primer pairs were used. These had been designed to amplify gene 3 (primer pairs PS1 + /PM4−, PS1 + /PM5−, PS3 + /PM4− and PS4 + /PM4 − ), gene 5 (primer pair PM2 + /PN2−, PM3 + /PN2 − ), and the 3′-untranslated region (UTR) (primer pair UTR41 + /UTR11 − ) (Cavanagh et al., Citation2001 Citation2002), and gene N (primer pair TVCnucleo + /TCVnucleo − ; Sellers et al., Citation2004) of avian (group 3) coronaviruses.
Figure 1. Relative positions of the pan-coronavirus oligonucleotide primers of Stephensen et al. (Citation1999), which are situated in the RNA-dependent RNA-polymerase encoding region of gene 1. The five-figure numbers in the smallest font size correspond to the nucleotide positions from the 5′ end of the IBV Beaudette genome. Dashed lines and associated numbers indicate the size of the PCR product (bp, base pairs) that would be generated with pairs of primers. nt, nucleotides.
The RT-PCR conditions were: RT step, 42°C for 30 min and 95°C for 5 min; followed by 35 cycles of 95°C for 1 min, 50°C for 1 min and 72°C for 1 min, with a final extension step of 72°C for 10 min. PCR products were analysed by agarose electrophoresis, sequenced, and sequences compared as described by Cavanagh et al. (Citation2001).
Phylogenetic analysis
The 66 amino acid sequences were aligned using ClustalX and analysed using the PHYLIP programs, PROTDIST using Kimura's distance method and NEIGHBOUR using randomization of the sequences.
Accession number
The partial sequence of the RdRp region of gene 1 of the parrot coronavirus has been submitted to GenBank and has been assigned the accession number DQ233651.
Results and Discussion
Virus isolation and electron microscopy
Following inoculation of primary cell cultures, a widespread CPE was seen in the macaw embryo liver cultures inoculated with samples of the liver, spleen and kidney. The CPE was characterized by the appearance of rounded, highly refractile cells that detached from the monolayer 2 to 3 days after inoculation. A similar CPE was noted in the CEL cultures, although two passages were required before the CPE was observed. Due to unavailability of macaw embryos, all subsequent propagation of the agent was undertaken in CEL cultures. No cytopathic agents were detected in the uninoculated control cultures following three blind passages. The inoculated SPF chicken embryos remained normal following three passages via the allantoic cavity and yolk sac.
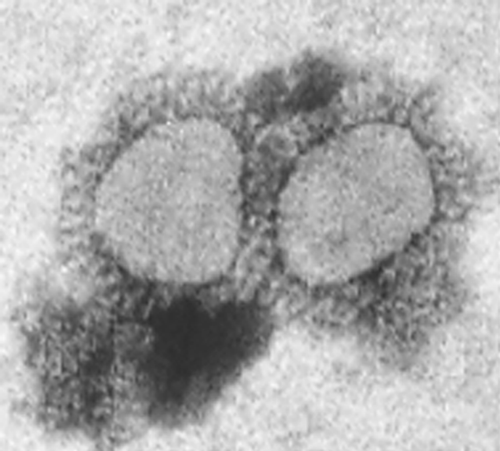
Negative contrast electron microscopy of material recovered from both the macaw embryo liver and CEL cultures following ultracentrifugation revealed spherical and pleomorphic virus particles, which in size and morphology resembled coronaviruses ().
RT-PCR and sequence analysis
The pan-coronavirus, degenerate oligonucleotide primers 8p, 1Ap and 2Bp (genome sense) and 4Bm and 7m (antigenome sense) correspond to sequences within the highly conserved RNA-dependent RNA polymerase-encoding region of gene 1b () (Stephensen et al., Citation1999).
Primer combinations 8p + /7m− and 1Ap + /7m− did not yield a PCR product with the parrot virus RNA. However, primer pair 2Bp + /4Bm− did yield a 251 base pair product, the size expected if the parrot virus was a coronavirus (data not shown). A continuous sequence of 66 amino acids was deduced (). BLAST analysis against the EBI database revealed identity with only one protein, namely the RdRp of coronaviruses. Comparison of the deduced amino acid sequence with that of coronaviruses representative of groups 1, 2, 3 and SARS-CoV revealed amino acid identity in the range 66 to 74% (). Within existing groups, coronavirus species have approximately 85% or more amino acid identity in this region. Sequence comparisons between strains of one group with those of another group reveal identities in the range 60 to 70%. These data confirmed the deduction made after the electron microscopy observations; the virus was a coronavirus—parrot coronavirus (PaCoV).
Figure 3. Comparison of the deduced amino acid sequences of the RdRp protein region corresponding to the parrot-derived sequence. The sequences were aligned using ClustalX and displayed using Genedoc. The comparison comprises selected species of coronaviruses representing each of the coronavirus groups, 1, 2 and 3. Where the sequences of more than one strain of a coronavirus species were available (e.g. MHV [murine hepatitis virus]), a representative sequence, identical to that of the other strains that are not included in the figure, has been used in this figure. The accession numbers of the sequences used are parrot coronavirus AV71/99 (accession number DQ233651), IBV Beau-CK (M94356), IBV LX4 (AY338732), TCoV VR911 (AF124991), pigeon coronavirus 03-653 (AJ854132), mallard duck coronavirus (AJ854130), greylag goose coronavirus 04-V48 (AJ854158), HCoV-NL63 (AY567487), PEDV CV777 (AF353511), bat coronavirus (AY864196), human coronavirus HCoV-229E (AF304460), porcine transmissible gastroenteritis virus TGEV Purdue (AJ011482), feline coronavirus (FcoV) UCD2 (AF124987), canine coronavirus (CcoV)1-71 (AF124986), MHV-A59 (AY700211), rat sialodacryoadenitis (RSDA) coronavirus (AF124990), bovine coronavirus (BcoV) Mebus strain (U00735), HCoV-OC43 (AF124989), haemagglutinating encephalomyelitis virus (HEV) strain 741 (AF124988) and SARS coronavirus, strain Urbani (AY278741).
![Figure 3. Comparison of the deduced amino acid sequences of the RdRp protein region corresponding to the parrot-derived sequence. The sequences were aligned using ClustalX and displayed using Genedoc. The comparison comprises selected species of coronaviruses representing each of the coronavirus groups, 1, 2 and 3. Where the sequences of more than one strain of a coronavirus species were available (e.g. MHV [murine hepatitis virus]), a representative sequence, identical to that of the other strains that are not included in the figure, has been used in this figure. The accession numbers of the sequences used are parrot coronavirus AV71/99 (accession number DQ233651), IBV Beau-CK (M94356), IBV LX4 (AY338732), TCoV VR911 (AF124991), pigeon coronavirus 03-653 (AJ854132), mallard duck coronavirus (AJ854130), greylag goose coronavirus 04-V48 (AJ854158), HCoV-NL63 (AY567487), PEDV CV777 (AF353511), bat coronavirus (AY864196), human coronavirus HCoV-229E (AF304460), porcine transmissible gastroenteritis virus TGEV Purdue (AJ011482), feline coronavirus (FcoV) UCD2 (AF124987), canine coronavirus (CcoV)1-71 (AF124986), MHV-A59 (AY700211), rat sialodacryoadenitis (RSDA) coronavirus (AF124990), bovine coronavirus (BcoV) Mebus strain (U00735), HCoV-OC43 (AF124989), haemagglutinating encephalomyelitis virus (HEV) strain 741 (AF124988) and SARS coronavirus, strain Urbani (AY278741).](/cms/asset/5102baa2-e823-45c3-af25-ed454058041e/cavp_a_159756_o_f0003g.gif)
Phylogenetic analysis of the 66-residue partial sequence of the RdRp of the PaCoV revealed that it did not coincide with any of the three existing coronavirus groups (). Although the RdRp is one of the most conserved protein sequences among coronaviruses, the small size of our sequence, only 66 residues, is such as to preclude at this stage any conclusions with regard to which, if any, of the existing groups the parrot virus might belong.
Figure 4. Phylogenetic relationship of the parrot-derived RdRp protein sequence compared with the corresponding region of the RdRp protein from coronaviruses representing each of the three groups. The 66 amino acid sequences were aligned using ClustalX and analysed using the PHYLIP programs, PROTDIST using Kimura's distance method and NEIGHBOUR using randomization of the sequences. The resulting tree is unrooted. The three coronavirus groups, 1 to 3, are highlighted. The accession numbers of the sequences are as indicated in .
Apart from the RdRp oligonucleotides, none of the other oligonucleotide pairs used in this study, based on IBV (group 3) sequences (see in Cavanagh et al., Citation2002), gave PCR products. These primer pairs had been chosen because they gave PCR products with coronaviruses from galliform (chicken-like) birds, 18 to 22 of 23 isolates of IBV, representing many serotypes collected over a 40-year period in several countries (unpublished observations), and also with turkey coronavirus (TcoV) and pheasant coronavirus (PhCoV) (Cavanagh et al., Citation2001 Citation2002; Sellers et al., Citation2004). Oligonucleotide pair UTR41 + /UTR11−, corresponding to parts of the 3′-UTR of IBV, gave PCR products with 23/23 strains of IBV, TCoV and PhCoV. This suggests a lower degree of relationship between the PaCoV and those of the galliform birds compared with the degree of identity exhibited among coronaviruses of galliform birds.
Serological analysis
Haemagglutination inhibition tests
Antiserum prepared in SPF chickens against the PaCoV was used in HI tests with HA antigen prepared with six serotypes of IBV and pheasant coronavirus. No inhibition of HA was observed. Repeated attempts to produce HA activity with the PaCoV-like agent by treatment with neuraminidase were unsuccessful.
Virus neutralization tests
VN tests in CEL cultures were carried out, but no significant heterologous neutralization was obtained with any of the reference IBV antisera or with the PaCoV antiserum. The PaCoV antiserum, which had a maximum homologous neutralizing titre of between 24 and 25, was also tested by VN test in embryonated SPF hens’ eggs using six reference strains of IBV, but no heterologous neutralization was obtained with any of the strains.
Agar gel precipitin test
The PaCoV antiserum was also tested using the IBV group-specific agar gel precipitation test but no lines of precipitation were detected.
The virion proteins of IBV are antigenically related to those of TCoV and PhCoV (Gough et al., Citation1996; Guy et al., Citation1997 Citation2002; Ismail et al., Citation2001). IBV exists as dozens of serotypes (Cavanagh, Citation2003; Liu & Kong, Citation2004; Cavanagh et al., Citation2005; Gelb et al., Citation2005), determined by the spike protein, so it was not surprising that there was no cross-reaction of the PaCoV serum with the six IBV serotypes and PhCoV in HI and virus neutralization tests. However, the N and M proteins are much more conserved among IBV, TCoV and PhCoV, resulting in antigenic cross-reactivity (Guy et al., Citation1997 Citation2002; Ismail et al., Citation2001; Chen et al., Citation2003). The lack of antigenic relationship between IBV and PaCoV revealed by the agar gel precipitation test indicates that the virion proteins of the parrot virus have substantially different sequences from those of the IBV-like viruses of group 3.
In a limited serological survey, 53 psittacine serum samples from a variety of sources, including four private aviaries, a collection in a zoological garden, quarantine aviaries and individual pet birds, were examined for neutralizing antibodies to the PaCoV. Antibodies to the isolate were detected in six macaw sera, with antibody titres ranging from 23 (1/8) to 26 (1/64). In three cases PDD had been reported in the aviaries from which positive sera had originated, but three other positive sera originated from birds in which PDD had not been reported.
Our investigation clearly shows that the virus isolated from the green-cheeked Amazon parrot with suspected PDD is a “new” coronavirus. Several coronaviruses have been detected in recent years in galliform birds—peafowl, Pavo cristatus (Liu et al., Citation2005); partridge, Alectoris sp.; and guinea fowl, Numida meleagris (Ito et al., Citation1991)—and non-galliform birds—teal, Anas sp. (Liu et al., Citation2005); greylag goose, Anser anser; mallard duck, Anas platyrhynchos; and pigeon, Columbia livia (Jonassen et al., Citation2005)—reviewed by Cavanagh (Citation2005). All of these viruses were group 3 viruses, with a high degree of nucleotide identity in their genes, including especially in the conserved region of the 3′-UTR and in the RdRp region of gene 1. The degree of amino acid identity between the 66-residue RdRp region of the parrot virus compared with coronaviruses of groups 1, 2 and 3 was similar to that obtained when species of one group are compared with those of another group, and less than that which exists between species of a given group. Further analysis of the PaCoV genome is required before it would be possible to state whether this virus should be considered a member of an existing coronavirus group or a new one.
It is unclear what the aetiological role of this coronavirus is in relation to PDD. The fact that the virus was isolated from a suspected case of PDD implies a causal relationship to the disease. During the course of this study a coronavirus-like agent was also isolated in CEL cultures from a Senegal parrot (Poicephalus senegalus) that had died in quarantine. Although macaw embryo liver cultures were unavailable for attempted virus isolation, the agent had a similar morphological appearance when examined by electron microscopy, replicated in CEL cell cultures causing a widespread CPE very similar to the PaCoV and appeared antigenically unrelated to IBV. In this case, however, there were no gross lesions of PDD and the cause of death was attributed to aspergillosis. Further transmission studies will be required to determine whether the parrot coronavirus of the present study is associated with PDD.
We believe this to be the first report of the isolation and characterization of a coronavirus from psittacine birds. In a previous study Hirai et al. (Citation1979) described the detection of a coronavirus-like agent in the livers of two Amazonia species of parrots. In contrast to our findings, these authors reported propagation of the isolates in embryonated fowls’ eggs but not in tissue cultures. However, subsequent studies by the same authors showed that the material containing the coronavirus-like agent was contaminated with Chlamydia psittaci, and when this organism was removed they were unable to demonstrate the embryo lethality associated with the coronavirus-like agent (Hirai et al., Citation1982). These authors were also unable to obtain a CPE with their coronavirus-like agent, unlike the coronavirus described in this report.
Acknowledgments
Thanks are due to Mark Evans MRCVS for referring the suspected case of PDD for investigation. Also to the Parrot Society (UK), Parrot Fund International and the Mid-West Avian Research Expo (MARE) for their financial support. Francesca Culver is a graduate student supported jointly by the British Poultry Council (Turkey Sector) and Merial Animal Health. Dave Cavanagh and Paul Britton are supported by the Department of the Environment, Food and Rural Affairs (grant OD0714), and by the Biotechnology and Biological Sciences Research Council.
References
- Cavanagh , D. 2003 . Severe acute respiratory syndrome vaccine development: experiences of vaccination against avian infectious bronchitis virus . Avian Pathology , 32 : 567 – 582 .
- Cavanagh , D. 2005 . Coronaviruses in poultry and other birds . Avian Pathology , 34 : 439 – 448 .
- Cavanagh , D. , Mawditt , K. , Sharma , M. , Drury , S.E. , Ainsworth , H.L. , Britton , P. and Gough , R.E. 2001 . Detection of a coronavirus from turkey poults in Europe genetically related to infectious bronchitis virus of chickens . Avian Pathology , 30 : 365 – 378 .
- Cavanagh , D. , Mawditt , K. , Welchman , D. , d , B , Britton , P. and Gough , R.E. 2002 . Coronaviruses from pheasants (Phasianus colchicus) are genetically closely related to coronaviruses of domestic fowl (infectious bronchitis virus) and turkeys . Avian Pathology , 31 : 181 – 193 .
- Cavanagh , D. , Picault , J.-P. , Gough , R.E. , Hess , M. , Mawditt , K. and Britton , P. 2005 . Variation in the spike protein of the 793/B type of infectious bronchitis virus, in the field and during alternate passage in chickens and embryonated eggs . Avian Pathology , 34 : 20 – 25 .
- Chen , H. , Coote , B , Attree , S. and Hiscox , J.A. 2003 . Evaluation of a nucleoprotein-based enzyme-linked immunosorbent assay for the detection of antibodies against infectious bronchitis virus . Avian Pathology , 32 : 519 – 526 .
- Gelb , J. Jr , Weisman , Y , Ladman , B.S. and Meir , R. 2005 . S1 gene characteristics and efficacy of vaccination against infectious bronchitis virus field isolates from the United States and Israel (1996 to 2000) . Avian Pathology , 34 : 194 – 203 .
- Gerlach , H. 1994 . “ Viruses ” . In Avian Medicine: Principles and Application , Edited by: Ritchie , B , Harrison , G. and Harrison , L. 862 – 948 . Lake Worth, FL : Wingers Publishing .
- Gough , R.E. , Alexander , D.J. , Collins , M.S. , Lister , S.A. and Cox , W.J. 1988 . Routine virus isolation or detection in the diagnosis of diseases of birds . Avian Pathology , 17 : 893 – 907 .
- Gough , R.E. , Cox , W.J. , Winkler , C.E. , Sharp , M.W. and Spackman , D. 1996 . Isolation and identification of infectious bronchitis virus from pheasants . The Veterinary Record , 138 : 208 – 209 .
- Guy , J.S. , Barnes , H.J. , Smith , L.G. and Breslin , J. 1997 . Antigenic characterization of a turkey coronavirus identified in poult enteritis and mortality syndrome-affected turkeys . Avian Diseases , 41 : 583 – 590 .
- Guy , J.S. , Smith , L.G. , Breslin , J.J. and Pakpinyo , S. 2002 . Development of a competitive enzyme-linked immunosorbent assay for detection of turkey coronavirus antibodies . Avian Diseases , 46 : 334 – 341 .
- Hirai , K. , Hitchner , S.B. and Calnek , B.W. 1979 . Characterisation of a new coronavirus-like agent isolated from parrots . Avian Diseases , 23 : 515 – 525 .
- Hirai , K. , Hitchner , S.B. and Calnek , B.W. 1982 . Correction in identification of a coronavirus-like agent isolated from parrots . Avian Diseases , 26 : 169 – 170 .
- Ismail , M.M. , Cho , K.O. , Hasoksuz , M. , Saif , L.J. and Saif , Y.M. 2001 . Antigenic and genomic relatedness of turkey-origin coronaviruses, bovine coronaviruses, and infectious bronchitis virus of chickens . Avian Diseases , 45 : 978 – 984 .
- Ito , N.M.K. , Miyaji , C.I. and Capellaro , C.E.M.P.D.M. 1991 . “ Studies on broiler's IBV and IB-like virus from guinea fowl ” . In II International Symposium on Infectious Bronchitis , Edited by: Kaleta , E.F. and Heffels-Redmann , U. 302 – 307 . Giessen : Justus Leibig University .
- Jonassen , C.M. , Kofstad , T. , Larsen , I.-L. , Lovland , A. , Handeland , K. , Follestad , A. and Lillehaug , A. 2005 . Molecular identification and characterization of novel coronaviruses infecting greylag geese (Anser anser), feral pigeons (Columba livia) and mallards (Anas platyrhynchos) . Journal of General Virology , 86 : 1597 – 1607 .
- Liu , S. and Kong , X. 2004 . A new genotype of nephropathogenic infectious bronchitis virus circulating in vaccinated and non-vaccinated flocks in China . Avian Pathology , 33 : 321 – 327 .
- Liu , S. , Chen , J. , Chen , J. , Kong , X. , Shao , Y. , Han , Z. , Feng , L. , Cai , X. , Gu , S. and Liu , M. 2005 . Isolation of avian infectious bronchitis coronavirus from domestic peafowl (Pavo cristatus) and teal (Anas) . Journal of General Virology , 86 : 719 – 725 .
- OIE (2004) . Avian infectious bronchitis . in Manual of Diagnostic Tests and Vaccines for Terrestrial Animals Chapter 2.7.6 . World Organisation for Animal Health (OIE) . Available online at : http://www.oie.int/eng/normes/mmanual/A_00107.htm ( Accessed 4 April 2005 .)
- Sellers , H.S. , Koci , M.D. , Linnemann , E. , Kelley , L.A. and Schultz-Cherry , S. 2004 . Development of a multiplex reverse transcription-polymerase chain reaction diagnostic test specific for turkey astrovirus and coronavirus . Avian Diseases , 48 : 531 – 539 .
- Stephensen , C.B. , Casebolt , D.B. and Gangopadhyay , N.N. 1999 . Phylogenetic analysis of a highly conserved region of the polymerase gene from 11 coronavirus and development of a consensus polymerase chain reaction assay . Virus Research , 60 : 181 – 189 .