Abstract
Five strains of infectious bronchitis virus isolated from commercial chickens from the state of Pennsylvania, USA during the years 1998 and 1999 were studied. The strains were selected for cross-challenge in specific pathogen free chickens and virus neutralization in chick embryos on the basis of partial S1 sequence amino acid identity values. The partial sequences analysed spanned the hypervariable amino terminus region of S1 from amino acid residues 48 to 219, based on the Beaudette strain. Using their S1 identity values, the strains represented a continuum of genetic, and thus antigenic, relationships. When compared with strain PA/5083/99, strain PA/Wolgemuth/98 had high sequence identity (96%) followed by PA/171/99 (85%), PA/5344/98 (70%) and PA/1220/98 (34%). The method of Archetti and Horsfall was used for calculating antigenic relatedness values of virus neutralization tests. The same formula was also applied to the percentage protection values of cross-challenge tests to derive protective relatedness values among the strains. The antigenic relatedness values, protective relatedness values, and the partial S1 sequence identity values were then analysed. The findings indicated partial S1 sequence identity values were more strongly correlated with protective relatedness values and than antigenic relatedness values.
La comparaison de la séquence du gène S1 du virus de la bronchite infectieuse est une meilleure méthode prédictive de l'immunité à l'épreuve chez les poussins que le sérotypage par séroneutralisation
Cinq souches de virus de la bronchite infectieuse isolées de poulets du commerce dans l'état de Pennsylvanie aux USA durant les années 1998 et 1999 ont été étudiées. Les souches ont été sélectionnées pour des épreuves croisées chez des poulets indemnes d'organismes pathogènes spécifiés et des séroneutralisations (VN) sur embryons de poulet sur la base des valeurs d'identité de la séquence partielle de S1 en acides aminés. Les séquences partielles analysées comprenaient les acides aminés 48 à 219 de la région hypervariable aminoterminale de S1 (numérotation basée sur la souche Beaudette). En utilisant les valeurs d'identité de S1, les souches représentaient un continuum de relations génétiques et donc antigéniques. En comparant les différentes souches à la souche PA/5083/99, la souche PA/Wolgemuth/98 présentait un identité de séquence élevée (90%) suivie par les souches PA/171/99 (85%), PA/5344/98 (70%) et PA/1220/98 (34%).
La méthode d'Archetti et Horsfall a été utilisée pour quantifier les niveaux de relation antigénique (ARV) dans les tests de VN. La même formule a également été appliquée aux valeurs de protection des tests d'épreuve croisée, exprimées en pourcentage, pour en déduire les niveaux de relation de protection (PRV) entre les souches. Les ARVs, PRVs et les valeurs d'identité de la séquence partielle de S1 ont été analysées. Les résultats ont montré que les valeurs d'identité de la séquence partielle de S1 présentaient une corrélation plus étroite avec les PRVS qu'avec les ARVs.
Der Vergleich der Sequenz des S1-Gens des Virus der infektiösen Bronchitis ermöglicht eine bessere Vorhersage der Belastbarkeit der Immunität in Hühnern als die Serotypisierung mittels Virusneutralisation
Es wurden fünf Stämme des Virus der infektiösen Bronchitis untersucht, die in den Jahren 1998 und 1999 im US-Staat Pennsylvania aus kommerziellen Hühnern isoliert worden waren.
Die Stämme wurden aufgrund partieller Aminosäurensequenzübereinstimmung des S1 für die Kreuzbelastungstests in spezifisch pathogen freien Hühnerküken und die Virusneutralisationstests in Hühnerembryonen ausgewählt. Die analysierten Teilsequenzen umfassten die hypervariablen Aminoterminusregion des S1 bezogen auf den Beaudette-Stamm von der Aminosäurenstelle 48 bis zur Aminosäure 219. Basierend auf ihren S1-Übereinstimmungswerten repräsentierten die Stämme ein Kontinuum von genetischen und somit auch antigenetischen Beziehungen. Im Vergleich mit dem Stamm PA/5083/99 wies der Stamm PA/Wolgemuth/98 die höchste Sequenzübereinstimmung (96 %) auf gefolgt von PA/171/99 (85 %), PA/5344/98 (70 %) und PA/1220/98 (34 %). Mittels der Methode nach Archetti und Horsfall wurden aus den Ergebnissen der Virusneutralisationstests die antigenetischen r-Werte (ARV) berechnet. Dieselbe Formel wurde auch verwendet, um aus den prozentualen Schutzwerten aus den Kreuzbelastungstests die schützenden r-Werte (PRV) zu bestimmen. Die ARVs, PRVs und Übereinstimmungswerte der S1-Teilsequenzen wurden dann analysiert. Die Ergebnisse ließen erkennen, das die Übereinstimmungswerte der S1-Teilsequenzen stärker mit den PRVS als mit den ARVs korrelierten.
La comparación de la secuencia del gen S1 del virus de la bronquitis infecciosa es un mejor indicador del desafío de la inmunidad en aves que la serotipificación mediante neutralización viral
Se estudiaron cinco cepas del virus de la bronquitis infecciosa aisladas en pollos comerciales del estado de Pensilvania, USA durante los años 1998 y 1999. Estas cepas se seleccionaron según los valores de similitud de las secuencias aminoacídicas parciales de la S1 para la neutralización viral (VN) y la infección cruzada en aves libres de patógenos específicos. Las secuencias parciales analizadas comprendían la región hipervariable amino terminal de la S1 desde la posición aminoacídica 48 a la 219, basado en la cepa Beaudette. En base a sus valores de similitud de la S1, las cepas representaban una continuación de las relaciones genéticas y por lo tanto antigénicas. Cuando se comparaba la cepa PA/5083/99, la secuencia de la cepa PA/Wolgemuth/98 tenía la mayor similitud (96%), seguida por PA/171/99 (85%), PA/5344/98 (70%) y PA/1220/98 (34%). Se utilizó el método de Archetti y Horsfall para calcular los valores de relación antigénica (Arv) de las pruebas de VN. También se aplicó la misma fórmula a los porcentajes de protección de las pruebas de infección-cruzada para obtener los valores de relación protectiva (PRV) entre las cepas. Entonces se analizaron los valores de similitud de la secuencia parcial de la S1, los ARVs y los PRVs. Estos hallazgos indicaron que los valores de similitud de la secuencia parcial de la S1 tenían una mayor correlación con PRVs que con los ARVs.
Introduction
Identification and antigenic characterization of infectious bronchitis virus (IBV) field strains often require one or more in vitro tests such as virus neutralization (VN) (Darbyshire et al., Citation1979; Wadey & Faragher, Citation1981), haemagglutination inhibition (King & Hopkins, Citation1984), monoclonal antibody reactivity (Koch et al., Citation1986; Karaca et al., Citation1992) and, more recently, analysis of sequences, commonly of the spike glycoprotein gene (Kusters et al., Citation1989; Cavanagh et al., Citation1998; Kingham et al., Citation2000). As valuable as the findings of these assays are, however, the major goal is to establish the relationship of field strains to vaccine strains using cross-challenge studies in the chicken (Cook et al., Citation1999). Vaccines are the primary method of controlling IBV infections in commercial chickens. The concept of classifying strains according to their “protectotype” has been proposed (Lohr, Citation1988; Dhinakar Raj & Jones, Citation1996). However, given the logistical limitations (facilities and costs) of performing cross-challenge tests in chickens, scientists must often rely on using in-vitro tests to establish the relationship of IBV field strains and vaccines.
The objectives of this study were to select and characterize strains of IBV that represented a continuum of genetic and antigenic relationships. The strains were compared by VN, cross-challenge in chickens, and analysis of partial S1 gene sequences. Using the method of Archetti & Horsfall (Citation1950), antigenic relatedness values (ARV) were calculated for VN findings and protective relatedness values (PRV) were calculated for cross-challenge results. The ARV and S1 amino acid identity values were then compared to determine which parameter provided the best correlation with PRV.
Materials and Methods
Chick embryos and chickens
Specific pathogen free (SPF) White Leghorn embryonated chicken eggs were obtained from Sunrise Farms (Catskill, New York, USA). Embryonated eggs were used for production of viral seed stocks, virus reisolation attempts and VN assays.
SPF White Leghorn chickens were purchased from Charles River SPAFAS Inc. (Norwich, Connecticut, USA). Chickens were used for cross-challenge experiments and antisera production. The chickens were housed in Horsfall isolator units. Feed and drinking water was provided ad libitum.
Viruses
Background information and Genbank accession numbers for the five IBV strains used in this study are presented in . Four viruses (PA/5083/99, PA/171/99, PA/5344/98, PA/1220/98) were obtained as allantoic fluid from the Poultry Diagnostic Laboratory, New Bolton Center, University of Pennsylvania, Kennett Square, PA, USA. The fifth strain, PA/Wolgemuth/98, is maintained as an allantoic fluid stock in our laboratory. PA/Wolgemuth/98 and PA/171/99 are nephropathogenic strains associated with an infectious bronchitis outbreak from 1998 to 2000 (Ziegler et al., Citation2002).
Table 1. IBV strains included in this study
Seed stocks were prepared (Gelb & Jackwood, Citation1998) and used at a low embryo passage number (less than eight passages). The stocks tested negative for avian adenoviruses by polymerase chain reaction (Xie et al., Citation1999), avian reoviruses by reverse transcriptase-polymerase chain reaction (Liu et al., Citation2003), and agglutination of chicken red blood cells.
Sequencing and analysis of the S1 gene
Extraction of IBV RNA, reverse transcriptase-polymerase chain reaction of S1 gene fragments and analysis of sequences were performed as described elsewhere (Gelb et al., Citation2005).
Virus neutralization
Reciprocal alpha VN tests were performed with each of the strains in SPF embryonated eggs (Thayer & Beard, Citation1998) using antisera prepared as described previously (Gelb & Jackwood, Citation1998). The endpoints for each of the VNs were determined (Reed & Muench, Citation1938). ARV were calculated for each strain (Archetti & Horsfall, Citation1950; Wadey & Faragher, Citation1981).
Reciprocal cross-challenge studies
Five cross-challenge studies were performed. Each trial used 66 SPF Leghorn chickens, 2 weeks old, assigned to six groups of 11 birds per group. In each study, chickens were immunized with a single strain using 104 to 105 median embryo infectious doses per bird via the intraocular route. Four weeks post immunization, 30 additional 6-week-old SPF chickens were obtained and assigned to each treatment group as challenge controls. The immunized and control birds were then challenged intraocularly with 104 to 105 median embryo infectious doses per chick of one of the five different challenge viruses. Five days post challenge, tracheal swabs were obtained for virus reisolation attempts in chicken embryos (Gelb et al., Citation1981). The percentage protection afforded by the immunizing strain against challenge with homologous and heterologous strains was calculated. PRV were calculated for each strain using the Archetti & Horsfall (Citation1950) equation. A cross-challenge index (CCI) was calculated by subtracting the percentage susceptible value of challenge control birds from the percentage susceptible value of cross-challenged birds. Percentage protection values of 0 were replaced with 0.01 to perform the calculations. The CCI is analogous to the neutralization index (NI) used in calculating the ARV. The CCIs were used to establish the r1 and r2 values, where r1 = CCI heterologous #2/CCI homologous #1, and r2 = CCI heterologous #1/homologous #2. The PRV was calculated by taking the square root of r1×r2 and multiplying by 100 to convert to a percentage.
Results
Sequencing and analysis of the S1 gene
Amino acid sequence identity values for the partial S1 sequences of the strains in the study are presented in . Sequence identity values established a continuum of relatedness to PA/5083/99; PA/Wolgemuth/98 (96% identity), PA/171/99 (85% identity), PA/5344/98 (70% identity) and PA/1220/98 (34% identity). These levels of S1 relatedness were considered relevant because they represent a wide range of identity values observed among IBV strains. presents an alignment of the five strains and locations of amino acid changes within the 180 residue S1 fragment of PA/Wolgemuth/98, PA/171/99, PA/5344/98 and PA/1220/98 compared with PA/5083/99. The number of amino acid substitutions within the hypervariable (HVR) and non-HVR regions of the fragment are presented in .
Figure 1. Comparison of the amino acid sequence of a 180-residue fragment of the S1 gene of IBV isolates PA/5083/99, PA/Wolgemuth/98, PA/171/99, PA/5344/98, and PA/1220/98. Residues differing from PA/5083/99 are shown. Amino acids identical to PA/5083/99 are shown by a dot (.), amino acid deletions are shown by a dash (–). The HVR as described by Wang et al. (Citation1994) is shaded. Non-shaded regions are considered non-HVR.
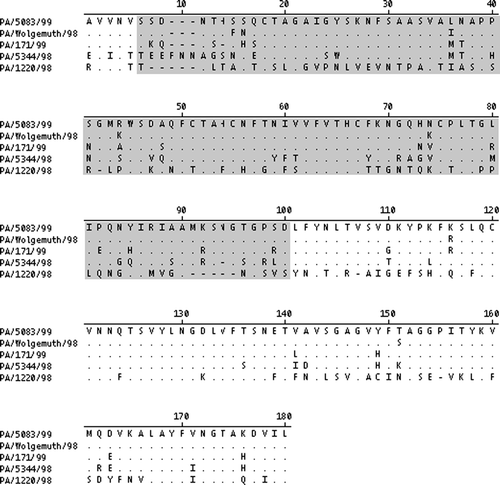
Table 2. Partial S1 amino acid sequence identity values (%) for Pennsylvania (PA) strains of IBV
Table 3. Amino acid changes in HVR and non-HVR regions of a 180-amino-acid fragment of the S1 gene of IBV using Pennsylvania (PA) strain PA/5083/99 as a basis of comparison
Virus neutralization
NI values from VN testing are presented in . By definition, NI values equal to or less than 2.0 represent no neutralization. NI values were used in the Archetti & Horsfall (Citation1950) equation to calculate the ARV presented in . Strains considered to be antigenically related have ARV of 50 to 100% (Wadey & Faragher, Citation1981). Antigenic relatedness among all of the isolates ranged from 4 to 67%. Strains PA/5083/99 and PA/Wolgemuth/98 were antigenically related (ARV = 67%). Strains PA/1220/98, PA/5344/98, and PA/171/99 were not antigenically related, as their ARV ranged from only 4 to 43%.
Table 4. Virus NI for homologous and heterologous reactions using Pennsylvania (PA) strains of IBV
Table 5. Antigenic relatedness values (%) based on virus NI of Pennsylvania (PA) strains of IBV
Reciprocal cross-challenge studies
Reciprocal cross-challenge findings are presented in . Challenge of chickens immunized with homologous strains produced protection ranging from 82 to 100%. Protection of immunized birds against challenge with heterologous strains ranged from 0 to 100%. All non-immunized challenge controls were susceptible to challenge (data not shown).
Table 6. Reciprocal cross-challenge findings performed in chickens using Pennsylvania (PA) strains of IBV
presents protective relatedness values derived from cross-challenge results for the five IBV strains.
Table 7. Protective relatedness values (%) based on reciprocal cross-challenge tests performed in chickens using Pennsylvania (PA) strains of IBV
Discussion
Assessing antigenic relationships among strains of IBV has been accomplished by several standard methods that include laboratory cross-challenge studies in chickens and VN and haemagglutination inhibition tests. The most comprehensive assessments have used reciprocal or two-way reactions to characterize the strains. This report describes the use of a PRV for the purpose of comparing levels of protection that are afforded to chickens immunized with one strain and later challenged with a heterologous strain.
In this study, protection was expressed as a PRV, derived from the Archetti & Horsfall (Citation1950) equation, which is also used to compare antigenic differences by VN. The PRV calculation utilizes the results of heterologous strain cross-challenge, homologous strain cross-challenge, and challenge of non-immunized controls. The PRV is a more accurate representation of the protective relatedness of two strains because of these internal controls that previously were often performed but were not included in a single value to represent strain similarities.
For heterologous strains that are highly related, the PRV may exceed 100, as is the case for strains PA/5083/99 and PA/Wolgemuth/98 (PRV = 104) (). This is not unprecedented however, when using the Archetti & Horsfall (Citation1950) equation for deriving an ARV. The PRV and ARV may be greater than 100 if heterologous strain test results for cross-protection or VN exceed values from homologous strain tests.
As observed previously and in this study, VN assessments of antigenic relationships between different strains generally are more discriminating than those based on challenge studies in chickens. Predictably, PRV based on cross-challenge studies were often greater than ARV based on reciprocal VN findings. The relationship between protection and serotype is often difficult to establish. IBV strains that have been shown to be of different serotypes by VN may be able to induce partial or complete immunity in chickens (Hitchner et al., Citation1964; Raggi & Lee, Citation1965; Winterfield et al., Citation1972; Darbyshire, Citation1980 Citation1985; Arvidson et al., Citation1990). The broader degree of cross-protection in birds is likely to be related to the cell-mediated immune responses to shared T-cell epitopes among some heterologous strains (Janse et al., Citation1994; Dhinakar Raj & Jones, Citation1997; Collisson et al., Citation2000; Seo et al., Citation2000).
PRV have practical implications for selecting strains for controlling IBV. Although not used in this study, vaccine strains could be included in cross-challenge studies to determine the PRV for a field strain associated with an outbreak.
Realistically, however, few IBV strains will be characterized by cross-challenge tests in chickens for the purposes of establishing PRV. Poultry biocontainment facilities needed for challenge studies are not widely available. Furthermore, some countries, including the USA, cannot easily obtain the permits needed to import live IBV field strains from other countries. Optimistically, however, it is hoped that field strains responsible for the most serious infectious bronchitis outbreaks would be used in challenge studies.
In-vitro assays, in most cases, will be used to identify field strains and the resulting findings will be used potentially to select a vaccine to control the disease. In this study, PRV from cross-challenge studies were found to have a higher degree of correlation with amino acid identity values derived from partial S1 gene analysis (0.72) than ARV calculated from VN tests (0.61) (). The S1 of IBV encodes epitopes important in VN (Cavanagh et al., Citation1992) and protective immunity (Cavanagh et al., Citation1997) in the chicken. Cavanagh et al. (Citation1997) examined the association of S1 sequence with protective immunity in their efforts to determine whether the S1 subunit was the major inducer of immunity in chickens. Differences in the challenge assessment method (ciliostasis in tracheal organ cultures versus tracheal IBV reisolation in embryos), the IBV strains used, and the specific amino acids included in the S1 analyses prevent direct comparison of their findings and ours. However, acknowledging the differences, both studies found that isolates with very high S1 sequence identities induced consistently higher levels of cross-protection than strains that had lower sequence identities. In most of their challenge trials, Cavanagh et al. (Citation1997) found that isolates highly related by S1 identities (98%) also induced a high degree of cross-protection. In our study, we also found that a very high PRV (104) and reciprocal cross-protection (100 and 91%) was conferred by strains PA/5083/99 and PA/Wolgemuth/98 with highly similar S1 identities (96%).
Table 8. Correlation between ARV, PRV and partial S1 amino acid sequence identity values of Pennsylvania (PA) strains of IBVa
PA/171/99 with 85% S1 sequence identities to both PA/5083/99 and PA/Wolgemuth/98, also induced relatively high cross-protection (PRV = 85 and PRV = 99, respectively). Again similar findings were reported by Cavanagh et al. (Citation1997) who showed that protection levels for heterologous challenges were only slightly lower than those of the homologous challenges using strains with 80% sequence identities. Moreover, both the Cavanagh et al. (Citation1997) study and our research also showed that strains with low S1 identities provide low or inconsistent cross-protection. Strains PA/5344/98 and PA/1220/98 with low S1 sequence identities (≤74%) to PA/5083/99, PA/Wolgemuth/98 and PA/171/99 resulted in low PRV (0.6 to 33%) in five of seven possible comparisons ( and ). However, somewhat higher PRV of 60 and 71% were observed, based primarily on one-way cross-challenge results for PA/5083/99 and PA/5344/98, as well as PA/171/99 and PA/1220/98. One-way cross-challenge protection has been reported previously (Hitchner et al., Citation1964; Hofstad, Citation1981).
Defining the most appropriate sequences used in the analysis of field strains and vaccines that provide the best prediction of protection is worthy of investigation. The partial S1 sequences used in the present study reflect a higher degree of variability between strains than, for example, would be the case if the entire S1 was analysed. These sequences were selected because they are routinely used in our laboratory for diagnostic analysis of field strains where turnaround time and costs must be considered. Had we used the sequences of the entire S1 gene, the correlation between PRV and sequencing would have been even higher—0.75 for full S1 compared with 0.72 for partial S1.
Strains with both highly similar and diverse S1 gene sequences were used in this study (). Amino acid substitutions in HVR and non-HVR regions of a 180-amino-acid residue S1 gene fragment were analysed ( and ). Consistent with other reports (Niesters et al., Citation1986; Cavanagh et al., Citation1988; Kusters et al., Citation1989; Kingham et al., Citation2000; Wang & Huang, Citation2000), amino acid substitutions were more numerous in the HVR than in the non-HVR. As expected, highly similar strains PA/5083/98, PA/Wolgemuth/98, and PA/171/99 had fewer overall amino acid changes than more diverse strains PA/5344/98 and PA/1220/98. Remarkably, PA/1220/98 had extensive variability in the non-HVR. The diversity of amino acids in the non-HVR of PA/1220/98 compared with all other strains used in this study may provide evidence for additional lineages within group III coronaviruses (IBV).
Using S1 identity values as a predictor of in-vivo protection would be a much more readily achievable alternative to performing laboratory challenge studies. In many cases, sequence analysis in the future will provide valuable practical information about the cross-protective potential between different strains. However, a word of caution is in order. Although not seen in this report, it is possible that some strains with high degrees of S1 identities may not cross-protect against challenge. A very few amino acid differences located in major immunodominant regions of the gene may be sufficient to cause a discrepancy between sequence and PRV correlations (Cavanagh et al., Citation1992).
Sequencing, although requiring the assistance of a specialized reference laboratory, is being used by an increasing number of regional and national laboratories in many countries. Laboratory research and clinical (field) application of its findings will ultimately establish the usefulness of S1 sequencing as a predictor of vaccine efficacy.
References
- Archetti , I. and Horsfall , F.L. 1950 . Persistent antigenic variation of influenza A viruses after incomplete neutralization in ovo with heterologous immune serum . Journal of Experimental Medicine , 92 : 441 – 462 .
- Arvidson , Y. , Tannock , G.A. , Senthilselvan , A. and Zerbes , M. 1990 . A model for determining immunogenic relationships between avian infectious bronchitis viruses . Archives of Virology , 111 : 227 – 238 .
- Cavanagh , D. , Davis , P.J. and Mockett , A.P.A. 1988 . Amino acids within hypervariable region 1 of avian coronavirus IBV (Massachusetts serotype) spike glycoprotein are associated with neutralization epitopes . Virus Research , 11 : 141 – 150 .
- Cavanagh , D. , Davis , P.J. , Cook , J.K.A. , Li , D. , Kant , A. and Koch , G. 1992 . Location of the amino acid differences in the S1 spike glycoprotein of closely related serotypes of infectious bronchitis virus . Avian Pathology , 21 : 33 – 43 .
- Cavanagh , D. , Ellis , M.M. and Cook , J.K.A. 1997 . Relationship between variation in the S1 spike protein of infectious bronchitis virus and the extent of cross-protection . Avian Pathology , 26 : 63 – 74 .
- Cavanagh , D. , Mawditt , K. , Adzhar , A. , Gough , R.E. , Picault , J.P. , Naylor , C.J. , Haydon , D. , Shaw , K. and Britton , P. 1998 . Does IBV change slowly despite the capacity of the spike protein to vary greatly? . Advances in Experimental Medicine and Biology , 440 : 729 – 734 .
- Collisson , E.W. , Pei , J. , Dzielawa , J. and Seo , S.H. 2000 . Cytotoxic T Lymphocytes are vital in the control of infectious bronchitis virus in poultry . Developmental and Comparative Immunology , 24 : 187 – 200 .
- Cook , J.K.A , Orbell , S.J. , Woods , M.A. and Huggins , M.B. 1999 . Breadth of protection of the respiratory tract provided by different live-attenuated infectious bronchitis vaccines against challenge with infectious bronchitis virus of heterologous serotypes . Avian Pathology , 28 : 477 – 485 .
- Darbyshire , J.H. 1980 . Assessment of cross-immunity in chickens to strains of avian infectious bronchitis virus using tracheal organ culture . Avian Pathology , 9 : 179 – 184 .
- Darbyshire , J.H. 1985 . A clearance test to assess protection in chickens vaccinated against infectious bronchitis virus . Avian Pathology , 14 : 497 – 508 .
- Darbyshire , J.H. , Rowell , J.G. , Cook , J.K.A. and Peters , R.W. 1979 . Taxonomic studies on strains of avian infectious bronchitis virus using neutralization tests in tracheal organ cultures . Archives of Virology , 61 : 227 – 238 .
- Dhinakar Raj , G. and Jones , R.C. 1996 . Protectotypic differentiation of avian infectious bronchitis viruses using an in vitro challenge model . Veterinary Microbiology , 53 : 239 – 252 .
- Dhinakar Raj , G. and Jones , R.C. 1997 . Effect of T-cell suppression by cyclosporin on primary and persistent infections of infectious bronchitis virus in chickens . Avian Pathology , 26 : 257 – 276 .
- Gelb , J. Jr and Jackwood , M.W. 1998 . “ Infectious bronchitis ” . In A Laboratory Manual for the Isolation and Identification of Avian Pathogens , 4th edn , Edited by: Swayne , D.E. , Glisson , J.R. , Jackwood , M.W. , Pearson , J.E. and Reed , W.M. 169 – 174 . Kennett Square, PA : American Association of Avian Pathologists .
- Gelb , J. Jr , Perkins , B.E. , Rosenberger , J.K. and Allen , P.H. 1981 . Serologic and cross-protection studies with several infectious bronchitis virus isolates from Delmarva-reared broiler chickens . Avian Diseases , 25 : 655 – 666 .
- Gelb , J. Jr , Weisman , Y. , Ladman , B.S. and Meir , R. 2005 . S1 gene characteristics and efficacy of vaccination against infectious bronchitis virus field isolates from the United States and Israel (1996–2000) . Avian Pathology , 34 : 194 – 203 .
- Higgins , D.G. and Sharp , P.M. 1988 . CLUSTAL: a package for performing multiple sequence alignment on a microcomputer . Gene , 73 : 237 – 244 .
- Hitchner , S.B. , Appleton , G.S. and Winterfield , R.W. 1964 . Evaluation of the immunity response to infectious bronchitis virus . Avian Diseases , 8 : 153 – 162 .
- Hofstad , M. S. 1981 . Cross-immunity in chickens using seven isolates of avian infectious bronchitis virus . Avian Diseases , 25 : 650 – 654 .
- Janse , M. , Van Roozelaar , D. and Koch , G. 1994 . Leukocyte subpopulations in kidney and trachea of chickens infected with infectious bronchitis virus . Avian Pathology , 23 : 513 – 523 .
- Karaca , K. , Naqi , S. and Gelb , J. Jr . 1992 . Production and characterization of monoclonal antibodies to three infectious bronchitis virus serotypes . Avian Diseases , 36 : 903 – 915 .
- Keeler , C.L. Jr , Reed , K.L. , Nix , W.A. and Gelb , J. Jr . 1998 . Serotype identification of avian infectious bronchitis virus by RT-PCR of the peplomer (S-1) gene . Avian Diseases , 42 : 275 – 284 .
- King , D.J. and Hopkins , S.R. 1984 . Rapid serotyping of infectious bronchitis virus isolates with the hemagglutination-inhibition test . Avian Diseases , 28 : 727 – 733 .
- Kingham , B.F. , Keeler , C.L. Jr , Nix , W.A. , Ladman , B.S. and Gelb , J. Jr . 2000 . Identification of avian infectious bronchitis virus by direct automated cycle sequencing of the S1 Gene . Avian Diseases , 44 : 325 – 335 .
- Koch , G. , Hartog , L. , Kant , A. , Van Roozelaar , D. and de Boer , G.F. 1986 . Antigenic differentiation of avian bronchitis virus variant strains employing monoclonal antibodies . Israeli Journal of Veterinary Medicine , 42 : 89 – 97 .
- Kusters , J.G. , Niesters , H.G. , Lenstra , J.A. , Horzinek , M.C. and van der Zeijst , B.A. 1989 . Phylogeny of antigenic variants of avian coronavirus IBV . Virology , 169 : 217 – 221 .
- Liu , H.J. , Lee , L.H. , Hsu , H.W. , Kuo , L.C. and Liao , M.H. 2003 . Molecular evolution of avian reovirus: evidence for genetic diversity and reassortment of the S-class genome segments and multiple cocirculating lineages . Virology , 314 : 336 – 349 .
- Lohr , J.E. (1988) . Differentiation of IBV strains . In E.F . Kaleta & U . Heffels-Redmann ( Eds .), Proceedings of the First International Symposium on Infectious Bronchitis (pp. 199 – 207 ). Rauischholzhausen, Germany .
- Niesters , H.G. , Lenstra , J.A. , Spaan , W.J. , Zijderveld , A.J. , Bleumink-Pluym , N.M. , Hong , F. , van Scharrenburg , G.J. , Horzinek , M.C. and van der Zeijst , B.A. 1986 . The peplomer protein sequence of the M41 strain of coronavirus IBV and its comparison with Beaudette strains . Virus Research , 5 : 253 – 263 .
- Raggi , L.G. and Lee , G.G. 1965 . Lack of correlation between infectivity, serological response and challenge results in immunisation with infectious bronchitis virus vaccine . Journal of Immunology , 94 : 538 – 543 .
- Reed , L.J. and Muench , H. 1938 . A simple method for estimating fifty percent endpoints . American Journal of Hygiene , 27 : 493 – 497 .
- Seo , S.H. , Pei , J. , Briles , W.E. , Dzielawa , J. and Collisson , E.W. 2000 . Adoptive transfer of infectious bronchitis virus primed alphabeta T cells bearing CD8 antigen protects chicks from acute infection . Virology , 30 : 183 – 189 .
- Thayer , S.G. and Beard , C.W. 1998 . “ Serologic procedures ” . In A Laboratory Manual for the Isolation and Identification of Avian Pathogens , 4th edn , Edited by: Swayne , D.E. , Glisson , J.R. , Jackwood , M.W. , Pearson , J.E. and Reed , W.M. 255 – 266 . Kennett Square, PA : American Association of Avian Pathologists .
- Wadey , C.N. and Faragher , J.T. 1981 . Australian infectious bronchitis viruses: identification of nine subtypes by a neutralization test . Research in Veterinary Science , 30 : 70 – 74 .
- Wang , L. , Junker , D. , Hock , L. , Ebiary , E. and Collisson , E.W. 1994 . Evolutionary implications of genetic variations in the S1 gene of infectious bronchitis virus . Virus Research , 34 : 327 – 338 .
- Wang , C.H. and Huang , Y.C. 2000 . Relationship between serotypes and genotypes based on the hypervariable region of the S1 gene of infectious bronchitis virus . Archives of Virology , 145 : 291 – 300 .
- Winterfield , R.W. , Fadly , A.M. and Bickford , A.A. 1972 . The immune response to infectious bronchitis virus determined by respiratory signs, virus infection, and histopathological lesions . Avian Diseases , 16 : 260 – 269 .
- Xie , Z. , Fadl , A.A. , Girshick , T. and Khan , M.I. 1999 . Detection of avian adenovirus by polymerase chain reaction . Avian Diseases , 43 : 98 – 105 .
- Ziegler , A.F. , Ladman , B.S. , Dunn , P.A. , Schneider , A. , Davison , S. , Miller , P.G. , Lu , H. , Weinstock , D. , Salem , M. , Eckroade , R. and Gelb , J. Jr . 2002 . Nephropathogenic infectious bronchitis in Pennsylvania chickens 1997–2000 . Avian Diseases , 46 : 847 – 858 .