Abstract
Following the avian influenza epidemics that occurred in Italy between 1997 and 2003, the Italian Ministry of Health in collaboration with veterinary authorities promoted, funded and implemented a national surveillance programme. The main objectives of the surveillance effort were to identify avian influenza viruses circulating in wild birds and to investigate the role of backyard poultry flocks in the dynamics of infection in a densely populated poultry area. Over 2 years (2004 to 2006), 164 backyard flocks and 4083 wild birds (mainly migratory Anseriformes and Charadriiformes) were sampled in three regions in the North of Italy. Samples collected were screened by means of real-time reverse transcriptase-polymerase chain reaction and the positive samples were processed for attempted virus isolation in embryonated fowl's specific pathogen free eggs. At the end of the study period, 27 low-pathogenic avian influenza viruses had been isolated from backyard flocks and 49 strains obtained from wild birds. Of these, 26 belonged to the H5 or H7 subtype and were closely related to contemporary low-pathogenic strains of Eurasian lineage. The findings confirm that backyard free-range farming is at high risk for avian influenza virus introduction, and confirm the role of wild waterfowl in the introduction and perpetuation of low-pathogenic avian influenza viruses during the winter season in Southern Europe.
Surveillance active des virus d'influenza aviaires chez les oiseaux sauvages et dans les troupeaux de volailles fermières dans le nord de l'Italie durant les années 2004–2006
Suite aux épisodes d'influenza aviaire (AI) qui ont sévi en Italie entre 1997 et 2003, le ministère italien de la santé en collaboration avec les autorités vétérinaires italiennes ont encouragé, financé et mis en place un programme de surveillance national. Les objectifs principaux de l'effort de surveillance ont été d'identifier les virus d'influenza aviaires (AIVs) circulant chez les oiseaux sauvages et d'investiguer le rôle des troupeaux de volailles fermières dans les dynamiques d'infection dans une zone à forte densité de population avicole (DPPA). Sur deux ans (2004–2006) 164 élevages fermiers et 4.083 oiseaux sauvages (principalement des ansériformes et des charadriiformes migrateurs) ont fait l'objet de prélèvements dans trois régions du nord de l'Italie. Les échantillons prélevés ont été étudiés par la réaction de transcription inverse suivie par la réaction de polymérisation en chaîne en temps réel (RRT-PCR); les échantillons positifs ont fait l'objet d'un essai d'isolement de virus sur œufs embryonnés de poules exemptes de microorganismes pathogènes spécifiés (SPF).
A la fin de l'étude, 27 virus d'influenza aviaires faiblement pathogènes (LPAI) ont été isolés à partir des élevages fermiers et 49 souches ont été isolées des oiseaux sauvages. Parmi ces isolats 26 appartenaient aux sous-types H5 ou H7 et étaient proches des souches faiblement pathogènes contemporaines du lignage Eurasien. Les résultats confirment que l'élevage fermier en plein air est exposé à un haut risque d'introduction des AIVs, et le rôle des oiseaux aquatiques sauvages dans l'introduction et la perpétuation des virus LPAI durant l'hiver dans le sud de l'Europe.
Aktive Überprüfung auf aviäre Influenzaviren bei Wildvögeln und Hinterhofherden in Norditalien in den Jahren 2004 bis 2006
Nach den aviären Influenza (AI)-Epidemien, die in Italien zwischen 1997 und 2003 aufgetreten sind, förderte, finanzierte und realisierte das italienische Gesundheitsministerium in Zusammenarbeit mit den Veterinärbehörden ein nationales Überwachungsprogramm. Hauptziel der Überwachungstätigkeit war die Identifizierung zirkulierender aviärer Influenzaviren (AIVs) bei Wildvögeln und die Untersuchung der Rolle von Hinterhofgeflügelherden in der Infektionsdynamik in einem dicht besiedelten Geflügelgebiet. Innerhalb von zwei Jahren (2004–2006) wurden in drei Regionen in Norditalien bei 164 Hinterhofherden und 4083 Wildvögeln (hauptsächlich Zugvögel aus den Ordnungen Anseriformes und Charadriiformes) Proben genommen. Die gesammelten Proben wurden mittels Real Time-Reverse Transkriptase-Polymerasekettenreaktion (RRT-PCR) überprüft und bei positiven Proben wurde versucht, das Virus in embryonierten spezifisch-pathogen-freien (SPF)-Hühnereiern anzuzüchten. Bis zum Ende der Studie konnten aus den Hinterhofherden 27 und den Wildvögeln 49 Stämme niedrig pathogener aviärer Influenza (LPAI)-Viren isoliert werden. Von diesen gehörten 26 Isolate zum H5- oder H7-Subtyp, wobei sie mit den aktuellen niedrig pathogenen Stämmen der eurasischen Linie eng verwandt waren. Diese Befunde belegen, dass die Freiland-Hinterhofhaltung ein großes Risiko für die Einschleppung von AIVs darstellt und dass wildlebendes Wassergeflügel bei der Einschleppung und Persistenz von LPAI-Viren in Südeuropa während der Wintersaison eine große Rolle spielt.
Vigilancia activa de virus de influenza aviar en aves salvajes y lotes de aves camperas en el Norte de Italia durante 2004–2006
Tras las epidemias de influenza aviar (AI) que ocurrieron en Italia entre 1997 y 2003, el Ministerio Italiano de Sanidad en colaboración con las autoridades veterinarias promovieron, financiaron y realizaron un programa de vigilancia nacional. El principal objetivo de esta vigilancia fue identificar virus de influenza aviares (AIVs) que estuvieran circulando en aves salvajes e investigar el papel que los lotes de aves camperas jugaban en la dinámica de la infección en las áreas de producción avícola de alta densidad (DPPA). Durante dos años (2004–1006) se muestrearon 164 lotes de aves camperas y 4083 aves salvajes (principalmente Anseriformes y Caradriformes migratorias) en tres regiones del Norte de Italia. Las muestras tomadas se procesaban mediante transcriptasa reversa-reacción en cadena de la polimerasa a tiempo real y en las muestras positivas se realizó aislamiento vírico en huevos embrionados de aves libres de patógenos específicos (SPF). Al final del estudio se habían aislado 27 virus de influenza aviar de baja patogenicidad (LPAI) a partir de lotes de aves camperas y 49 cepas de aves salvajes. De éstas, 26 pertenecían a los subtipos H5 o H7 y estaban estrechamente relacionados con cepas contemporáneas de la línea Euroasiática. Estos hallazgos confirman que las aves camperas criadas en libertad suponen un riesgo alto de introducción de AIVs, y el papel que las aves salvajes acuáticas juegan en la introducción y perpetuación de virus LPAI durante el invierno en el sur de Europa.
Introduction
Prior to the ongoing H5N1 avian influenza (AI) panzootic, Italy had been affected by several incursions of AI viruses of the H5 and H7 subtypes (Capua et al., Citation1999; Capua & Marangon, Citation2000; Capua et al., Citation2003). The consequences of these outbreaks were very severe for the poultry industry, and for this reason Italian veterinary authorities implemented a nationwide surveillance programme beginning in 2004.
This programme is still ongoing, and has revealed itself useful in understanding better the complex ecology of avian influenza viruses (AIVs) and has had a positive effect on the public opinion. In particular, such efforts are perceived as actions taken by public health authorities against the westward spread of the Asian H5N1 virus (Gilbert et al., Citation2006; Munster et al., Citation2006; Olsen et al., Citation2006).
The main objectives of the surveillance effort were to identify any AIVs circulating in wild birds and to establish whether these viruses were also infecting backyard poultry, which are thought to act as a link between the wild host and intensively reared poultry. The surveillance effort was focused in North Eastern Italy, as this is where AI epidemics had occurred in the past, and, incidentally, the area has a unique combination of densely populated poultry areas (DPPAs) and wetlands.
The wetlands are located along the Eurasian Anatidae flyway, and therefore represent resting and wintering sites for many wild ducks and geese migrating south from Northern and Eastern Europe. In addition, waders migrating south stop to rest and feed in the same wetlands, before undertaking the long flight over the Mediterranean Sea that precedes their over-wintering in Africa. The migrant population includes a high proportion of juveniles, hatched during the previous spring. Young birds are known to be highly susceptible to influenza virus infection (Hinshaw et al., Citation1980). Therefore, during the winter season, thousands of susceptible migratory birds originating from different sites in Northern and Eastern Europe congregate in Italian wetlands and intermingle. This situation represents an ideal ecological setting for the transmission and perpetuation of AIVs.
Materials and Methods
Specimens from backyard flocks
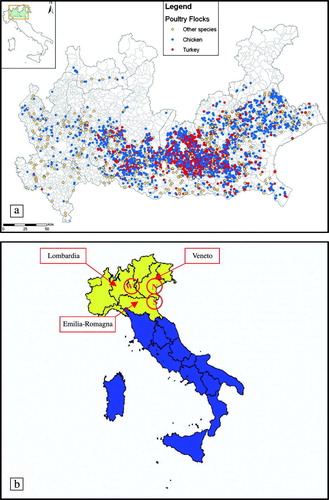
Between September 2004 and March 2006 a total of 164 backyard flocks in three regions, Lombardia, Veneto and Emilia-Romagna, were selected in areas at risk of infection. The number and location of backyard flocks to be sampled were identified by regional veterinary epidemiological centres on the basis of risk factors such as proximity of the farm with wetlands or DPPA and rearing of mixed species in the open. The sampling scheme was developed with the criterion that in case of infection at least one positive farm would be detected taking into account an estimated prevalence of 5% with a confidence level of 95%.
All selected backyard flocks raised primarily waterfowl. Commonly reared species were: mallard (Anas platyrhynchos), Pekin duck (A. platyrhynchos f. dom.), muscovy duck (Cairina moschata) and goose (Anser anser f. dom.). Often, Galliformes such as chickens (Gallus gallus), turkeys (Meleagris gallopavo) and guinea fowl (Numida meleagridis) were also present in the same flock. The surveillance programme was designed to allow monitoring of the same farm approximately every 45 days. In these flocks a minimum of 10 cloacal swabs from each representative species (20 in farms with more than 250 birds, or from all birds if less than 10), and at least one sample of fresh dropping from ducks and/or geese were collected for AI detection. The cloacal swabs (pool of five samples) and faecal samples (single or pooled) were analysed directly by virus isolation in embryonated chicken eggs without performing any preliminary screening tests.
Specimens from wild birds
During the winter seasons 2004/2005 and 2005/2006, 4083 cloacal swabs from wild birds were collected from 38 different species present in different wetlands of Northern Italy (Lagoon of Venice, Salt marsh of Cervia in Emilia-Romagna region, and the Po river delta). The samples were collected from trapped birds in bird-ringing stations or from birds shot during normal hunting activities.
The species sampled were mainly migratory Anseriformes and Charadriiformes () that fly into Italy from North and East European countries (a and 2b). For the sake of clarity, the main collection sites have been named North (N), South (S) and West (W) sampling areas (b) according to their geographic location in the study area. The collection sites include the most important wintering areas in North Italy. The geographical vicinity of the collection sites to the DPPA in Northern Italy is presented in a and 1b.
Figure 2. European flyways of (2a) mallard and (2b) common teal (Chelini, Citation1984).
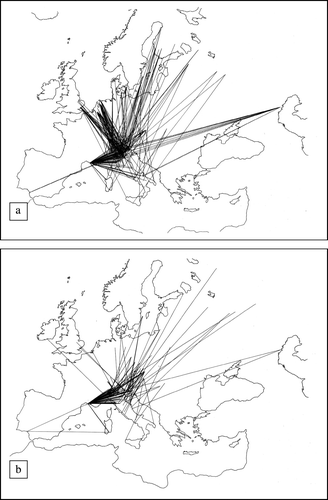
Table 1. Main wild bird species (≥10 subjects) tested during the study period: results of real-time RRT-PCR analyses and virus isolation
Cloacal swabs were collected by trained personnel (gamekeepers, bird census takers, ornithologists, veterinarians) with cotton swabs and were stored in transport media composed of phosphate-buffered saline/glycerol (9/1) containing antibiotics (10 000 U/ml penicillin, 10 mg/ml streptomycin, 0.25 mg/ml gentamycin and 5000 U/ml mycostatin). These samples were taken to the diagnostic laboratory within 24 to 48 h in a refrigerated container (+2 to +8°C) and stored at −80°C until analysed.
RNA detection and virus isolation
The samples collected from wild birds were screened by means of real-time reverse transcriptase-polymerase chain reaction (RT-PCR) specific for type A influenza viruses (Spackman et al., Citation2002; Cattoli et al., Citation2004). Viral RNA was extracted from allantoic fluid using the High Pure TM RNA Isolation Kit (Roche®). Samples with threshold cycle value ≤35 were considered positive for influenza A viral RNA and were subsequently processed for virus isolation using standard methods. Briefly, 100 µl original sample were inoculated into the allantoic cavity of 9-day-old to 11-day-old embryonating specific pathogen free eggs for virus isolation attempts according to EU Directive 92/40 (CEC, Citation1992). Haemagglutinating isolates were characterized by means of the haemagglutination inhibition tests and neuraminidase inhibition tests using specific hyperimmune chicken antisera to the reference strains of influenza virus (Alexander & Spackman, Citation1981). Isolates belonging to the H5 and H7 subtypes were pathotyped by sequencing the haemagglutinin gene segment (Hoffman et al., Citation2001). Sequences were obtained using the BigDye Terminator v 3.1 cycle sequencing kit (Applied Biosystems) in a 3100-Avant Genetic Analyzer (Applied Biosystems).
The nucleotide sequences obtained were aligned by Clustal W, and the evolutionary distances were calculated by Kimura's two-parameter method (Nei & Kumar, Citation2000). Phylogenetic dendrograms were constructed using the Neighbor Joining method and bootstrap analysis (1000 replicates) by MEGA 3 software (Nei & Kumar, Citation2000; Kumar et al., Citation2004). Branches with bootstrapping values ≥70 were considered significant, corresponding to a confidence interval ≥95% (Hillis & Bull, Citation1993).
Accession numbers
The sequences of the haemagglutinin gene segments of representative H5/H7 strains have been deposited in GenBank with the following accession numbers: A/mallard/Italy/3401/05 (H5N1), accession number DQ838508; A/goose/Italy/4692-11/04 (H7N7), accession number DQ838511; A/mallard/Italy/4234/04 (H7N7), accession number DQ838510; A/teal/Italy/3812/05 (H5N3), accession number DQ838509; A/duck/Italy/4692-30/04 (H7N7), accession number DQ838512; A/mallard/Italy/4776-36/04 (H7N7), accession number DQ838513; A/mallard/Italy/4810-7/04 (H7N7), accession number DQ838514; A/mallard/Italy/4810-79/04 (H7N4), accession number DQ838515; and A/teal/Italy/3931/2005 (H5N2), accession number DQ851561.
Results
Over the study period, 27 AIVs were isolated from 19 of 164 backyard flocks tested (11.6%). Apart from one virus isolated from a guinea fowl, all isolates were obtained from domestic Anseriformes (). Among the isolates we obtained only three viruses belonging to H5 or H7 subtype; namely, an H5N3 virus of low-pathogenicity avian influenza (LPAI) and two H7N7 LPAI isolates.
Table 2. Avian influenza isolates in Italy in 2004/2005 in backyard flocks
The majority of viruses isolated (20/27) from backyard flocks were obtained from samples collected during the autumn/winter season. Only seven positive samples were collected in the spring/summer season.
Of the 4083 wild birds tested, 327 were positive for influenza type A by real-time RT-PCR (). Following virus isolation attempts, 47 AIVs belonging to 15 different subtype combinations (H1N1, H1N3, H3N8, H4N6, H5N1, H5N2, H5N3, H6N2, H7N4, H7N7, H9N2, H10N4, H10N7, H10N8, H11N9) were obtained from migratory waterfowl belonging to the Order Anseriformes (). One AIV of the H10N7 subtype was isolated from a wintering common snipe (Gallinago gallinago) in a hunting territory located in the western part of the study area. This appears to be the first isolation of an AIV from a wader in Italy. Another unexpected finding was the isolation of a virus of H6N2 subtype from a greater flamingo (Phoenicopterus roseus).
Table 3. AI isolates from wild birds in each region in the winter seasons 2004/2005 and 2005/2006
No highly pathogenic avian influenza (HPAI) viruses were detected.
Phylogenetic dendrograms were constructed for viral isolates of the H7 and H5 subtypes in order to assess their relationship with the viruses of the same subtypes causing outbreaks in domestic poultry in Europe in the recent past ( and ). With reference to sequence and phylogenetic analysis of the haemagglutinin gene of H7 viruses isolated from wild birds, two separate sublineages could be identified. These formed a larger cluster with the H7N3 viruses causing the recent LPAI outbreak in Italy.
Figure 3. Phylogenetic analysis of the genome segment encoding for the H7 strains isolated from domestic and wild birds. Note: the strains in the oval (A/duck/Italy/4692-30/04, A/goose/Italy/4692-11/04 and A/mallard/Italy/4776-30/04), isolated respectively from domestic ducks and geese and from wild mallard in the 2004/2005 winter season cluster, cluster together.
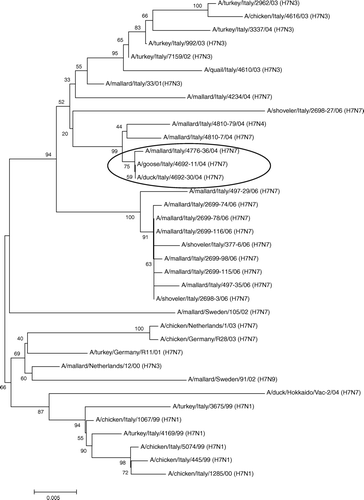
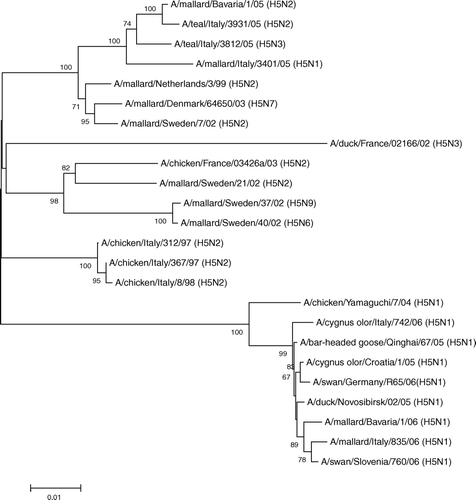
Figure 4. Phylogenetic analysis of the genome segment encoding for the H5 strains isolated from domestic and wild birds.
It is interesting to point out that H7 isolates collected in domestic and wild waterfowl at the end of 2004 (see strains in the oval in ) are closely related showing a high homology at the nucleotide level (up to 99.3%).
The H7 isolates from wild birds were quite distinct from the H7 viruses responsible for the last two severe HPAI outbreaks in Europe (homology <96.8% with the Italian strain H7N1 and the Dutch strain H7N7).
Similarly, the H5 viruses isolated from wild birds in 2005 clustered together in a group that is clearly distinct from the HPAI H5 viruses that caused outbreaks in 1997 and 1998 in domestic poultry in Italy and the contemporary HPAI H5N1 circulating in wild birds in Europe, Africa and Asia (). This clustering was supported by a high bootstrap value (100).
The LPAI H5 viruses were instead closely related to LPAI isolates from wild ducks sampled recently in North and Central European countries, in particular with the German strain A/mallard/Bavaria/1/2005 (H5N2) (nucleotide homology >98%).
Discussion
This study confirms previous data (Campitelli et al., Citation2004; De Marco et al., Citation2005; Terregino et al., Citation2005) indicating the active circulation of influenza A viruses in wild waterfowl in Italy, and highlights the role of wild and domestic waterfowl in the perpetuation of LPAI in nature (Webster & Hulse, Citation2004). In detail, certain species of wild waterfowl such as mallards, teals, pintails and wigeons should be considered as targets for sampling since they have been shown to harbour the majority of AIVs obtained during this study. The greater susceptibility of these species to AI infection has also been reported in other studies (Olsen et al., Citation2006).
The 49 strains isolated from wildfowl indicate that Italian wetlands represent, particularly during winter, an ecosystem that is able to perpetuate AIVs efficiently. The geographical vicinity between the wetlands and DPPAs suggest that they represent a risk for industrial poultry.
In comparing these data with others collected in Europe, it appears that the real-time RT-PCR method used in our study is more sensitive than other systems such as RT-PCR. In fact, in our study 15% of total samples were positive in real-time RT-PCR while in other studies a prevalence ranging from 1% to 2.3% were detected by RT-PCR (Fouchier et al., Citation2003; Munster et al., Citation2005).
However, given the low isolation rate (approximately 15%) of real-time RT-PCR positives and the overall virus isolation rates of AIVs from wild birds, it may seem reasonable to improve the outcome of isolation attempts. Possibly the quality of the specimen or the transportation medium or storage temperature and conditions could be improved. However, an additional possible explanation for the paucity of virus isolates obtained could be related to the level and duration of viral shedding in these birds once they arrive in Italy. It appears that influenza virus detection can be very high in Northern Europe prior to the winter migration (Okazaki et al., Citation2000; Olsen et al., Citation2006), and it is possible that a progressive decrease of shedding occurs during the southbound migration (Guberti et al., Citation2007). Birds that migrate from Northern and Eastern Europe congregate in Central Europe for several weeks before commencing the final stage of their southbound migration. During this period, birds may experience multiple infections with different subtypes, and would most probably develop an immunity to the most prevalent strains. An increasing level of population immunity would most probably lead to a progressive decrease in the levels of infection. For this reason it is possible that the levels of virus shedding in the majority of birds that reach Italy are influenced by the time interval between infection and sampling, by the distance covered and by the level of population immunity to prevalent subtypes. The perpetuation of the endemic cycle of AI under these conditions can probably only be guaranteed by the great numbers of birds present at one time in the wetlands (Delogu et al., Citation2003).
A noteworthy finding of this investigation is the difference in the isolation rates of H5 and H7 viruses over the 2-year period. During 2004/2005 there appeared to be a greater isolation rate of H7 viruses (8/11) and no H5 viruses were detected. In contrast during 2005/2006 the prevalence of H5 (3/37) increased and the prevalence of the isolates typed as H7 subtype (13/37) decreased. The reason for this remains to be clarified.
The present investigation also includes the first isolation of an AIV from a wader (common snipe) in Italy. These birds are thought to play a significant role in AI epidemiology in North America (Stallknecht & Shane, Citation1988), while in Europe their role is still to be fully understood. Another unusual finding is the isolation of an AIV from a great flamingo. These African birds have started colonizing selected areas of Italian wetlands since the 1990s (Sardinia, Apulia, Emilia-Romagna). Recently they have also colonized areas in Veneto and Sicily. Although their migratory behaviour is still under study, they are now considered semi-resident birds. Their presence in areas that are colonized by infected waterfowl may have unpredictable eco-epidemiological implications. In this study we collected 678 samples from charadriiform birds (waders, terns and gulls), and of these only one yielded a viable isolate. This rate of isolation success is much lower (less than 0.15% of all collected samples) than in the Order Anseriformes (approximately 1.5%).
The reason behind the low isolation rate from samples collected from Charadriiformes remains unclear. However, real-time RT-PCR positive samples collected from Charadriiformes were proportionally less than those collected from Anseriformes.
The findings in backyard flocks indicate that this type of farming is at high risk for AI introduction. This is primarily due to the fact that these holdings are mostly free range. The availability of food attracts wild birds and results in intermingling and in the deposition of droppings. All these conditions favour the introduction of AI into domesticated birds.
The isolation of two virtually identical H7N7 isolates () from a wild mallard and domestic waterfowl confirms this thesis. The risk of introduction of AIVs into free-range farms becomes very high during the period in which the highest number of natural reservoirs is present in the wetlands (autumn and winter). This is supported by the high isolation rate of AI viruses (74%) in backyard flocks during the colder months.
Penetration of AIVs into intensively farmed poultry may occur following amplification of infection in the domestic reservoir. It is also possible that, through trade or movement of backyard birds or of contaminated organic material, subsequent infection may spread significantly, thus representing an increased risk for intensively reared birds.
Regardless of the size of the flock, the role of free-range birds in acting as index cases of AIV introduction from the wild host has been highlighted in recent HPAI outbreaks occurring in Europe (Romania, Sweden, Germany, Denmark) (Euro Surveillance, Citation2006a,Citationb). Given the increased public perception of AI infections and the animal and human health issues, the issue of protecting free-range birds from infection should be addressed in a harmonized manner across the European Union.
The wealth of isolates that are now available across Europe and in other parts of the world, due to increased surveillance efforts, should be perceived and managed as a unique opportunity to increase our knowledge on AI and thus contribute to improve animal and public health. The degree of reassortment that is occurring in the wild bird population and the exchange of genes between viruses originating from different hosts should be investigated, along with the genetic variations that occur as AIVs infect new ecosystems and hosts. It is imperative that information collected in national efforts should be seen as part of a pan-European scenario and collated in order to maximize the practical outcome of such programmes.
Acknowledgements
The authors gratefully acknowledge all the staff of the Virology and Research and Development laboratories for their invaluable technical assistance, and Dr Mauro Delogu of the University of Bologna and Dr Giuseppe Barigazzi of the Department of Parma of IZSLER who supplied the Italian LPAI H5N1. This research was funded in part by the Italian Minister of Health (RC IZSVE 003/2001) and the Veneto Region (DGR 3398/05).
References
- Alexander , D.J. and Spackman , D. 1981 . Characterisation of influenza A viruses isolated from turkeys in England during March–May 1979 . Avian Pathology , 10 : 281 – 293 .
- Campitelli , L. , Mogavero , E. , De Marco , M.A. , Delogu , M. , Puzelli , S. , Frezza , F. , Facchini , M. , Chiapponi , C. , Foni , E. , Cordioli , P. , Webby , R. , Barigazzi , G. , Webster , R.G. and Donatelli , I. 2004 . Interspecies transmission of an H7N3 influenza virus from wild birds to intensively reared domestic poultry in Italy . Virology , 323 : 24 – 26 .
- Capua , I. and Marangon , S. 2000 . The avian influenza epidemic in Italy, 1999–2000: a review . Avian Pathology , 29 : 289 – 294 .
- Capua , I. , Marangon , S. and Cancellotti , F.M. 2003 . The 1999–2000 avian influenza (H7N1) epidemic in Italy . Veterinary Research Communication , 27 : 123 – 127 .
- Capua , I. , Marangon , S. , Selli , L. , Alexander , D.J. , Swayne , D.E. , Dalla Pozza , M. , Parenti , E. and Cancellotti , F.M. 1999 . Outbreaks of highly pathogenic avian influenza (H5N2) in Italy during October 1997 to January 1998 . Avian Pathology , 28 : 455 – 460 .
- Cattoli , G. , Drago , A. , Maniero , S. , Toffan , A. , Bertoli , E. , Fassina , S. , Terregino , C. , Robbi , C. , Vicenzoni , G. and Capua , I. 2004 . Comparison of three rapid detection systems for type A influenza virus on tracheal swabs of experimentally and naturally infected birds . Avian Pathology , 33 : 432 – 437 .
- CEC ( 1992 ). Council Directive 92/40/EEC of 19 May 1992 introducing Community measures for the control of avian influenza . Official Journal of the European Commission , L167 , 1 – 15 .
- Chelini , A. 1984 . Le anatre selvatiche , Firenze : Editoriale Olimpia .
- De Marco , M.A. , Foni , E. , Campitelli , L. , Delogu , M. , Raffini , E. , Chiapponi , C. , Barigazzi , G. , Cordioli , P. , Di Trani , L. and Donatelli , I. 2005 . Influenza virus circulation in wild aquatic birds in Italy during H5N2 and H7N1 poultry epidemic periods (1998 to 2000) . Avian Pathology , 34 ( 6 ) : 480 – 485 .
- Delogu , M. , De Marco , M.A. , Donatelli , I. , Campitelli , L. and Catelli , E. 2003 . Ecological aspects of influenza A virus circulation in wild birds of the western paleartic . Veterinary Research Communications , 27 : 101 – 106 .
- Euro Surveillance . ( 2006a , April 6 ). Avian influenza H5N1 detected in German poultry and a United Kingdom wild bird . Euro Surveillance , 11(4), E060406.1. Available online at: http://www.eurosurveillance.org/ew/2006/060406.asp
- Euro Surveillance . ( 2006b , May 25 ). Avian influenza H5N1 outbreaks in Romanian and Danish poultry, and large H5N1 cluster in an Indonesian family . Euro Surveillance , 11(5), E060525.1. Available online at: http://www.eurosurveillance.org/ew/2006/060525.asp
- Fouchier , R.A.M. , Olsen , B. , Bestebroer , T.M. , Herfst , S. , van der Kemp , L. , Rimmelzwaan , G.F. and Osterhaus , A.D.M.E. 2003 . Influenza A virus surveillance in wild birds in Northern Europe in 1999 and 2000 . Avian Diseases , 47 : 857 – 860 .
- Gilbert , M. , Xiao , X. , Domenech , J. , Lubroth , J. , Martin , V. and Slingenbergh , J. 2006 . Anatidae migration in the Western Palearctic and spread of highly pathogenic avian influenza H5N1 virus . Emerging Infectious Diseases , 12 ( 11 ) : 1650 – 1656 .
- Guberti , V. , Scremin , M. , Busani , L. , Bonfanti , L. and Terregino , C. 2007 . A simulation model for the LPAI viruses in dabbling ducks in Europe . Avian Diseases , 50 : 275 – 278 .
- Hillis , D.M. and Bull , J.J. 1993 . An empirical test of bootstrapping as a method for assessing confidence in phylogenetic analysis . Systematic Biology , 42 ( 2 ) : 182 – 192 .
- Hinshaw , V.S. , Webster , R.G. and Turner , B. 1980 . The perpetuation of Orthomyxoviruses and paramyxoviruses in Canadian waterfowl . Canadian Journal of Microbiology , 26 : 622 – 629 .
- Hoffman , E. , Stech , J. , Guan , Y. , Webster , R.G. and Perez , D.R. 2001 . Universal primer set for the full-leght amplification of all influenza A viruses . Archives of Virology , 146 : 2275 – 2289 .
- Kumar , S. , Tamura , K. and Nei , M. 2004 . MEGA 3: integrated software for molecular evolutionary genetics analysis and sequence alignment . Briefings in Bioinformatics , 5 ( 2 ) : 150 – 163 .
- Munster , V. , Wallensten , A. , Olsen , B. , Rimmelzwaan , G.F. , Osterhaus , A.D.M.E. and Fouchier , R.A.M. 2005 . “ Influenza A virus surveillance in wild birds ” . In Avian Influenza: Prevention and Control , Edited by: Schrijver& , R.S. and Koch , G. 25 – 30 . Dordrecht , , The Netherlands : Springer .
- Munster , V.J. , Veen , J. , Olsen , B. , Vogel , R. , Osterhaus , A.D.M.E. and Fouchier , R.A.M. 2006 . Towards improved influenza A virus surveillance in migrating birds . Vaccine , 24 ( 44–46 ) : 6729 – 6733 .
- Nei , M. and Kumar , S. 2000 . Molecular Evolution and Phylogenetics , 37 – 38 . New York : Oxford University Press, Inc .
- Okazaki , K. , Takada , A. , Ito , T. , Imai , M. , Takakuwa , H. , Hatta , M. , Ozaki , H. , Tanizaki , T. , Nagano , T. , Ninomiya , A. , Demenev , V.A. , Tyaptirganov , M.M. , Karatayeva , T.D. , Yamnikova , S.S. , Lvov , D.K. and Kida , H. 2000 . Precursor genes of future pandemic influenza viruses are perpetuated in ducks nesting in Siberia . Archives of Virology , 145 ( 5 ) : 885 – 893 .
- Olsen , B. , Munster , V.J. , Wallensten , A. , Waldenstrom , J. , Osterhaus , A.D. and Fouchier , R.A. 2006 . Global patterns of influenza a virus in wild birds . Science , 312 ( 5772 ) : 384 – 388 .
- Spackman , E. , Senne , D.A. , Myers , T.J. , Bulaga , L.L. , Garber , L.P. , Perdue , M.L. , Lohman , K. , Daum , L.T. and Suarez , D.L. 2002 . Development of a real-time reverse transcriptase PCR assay for type A influenza virus and the avian H5 and H7 hemagglutinin subtypes . Journal of Clinical Microbiology , 40 : 3256 – 3260 .
- Stallknecht , D.E. and Shane , M. 1988 . Host range of avian influenza virus in free-living birds . Veterinary Research Communications , 12 : 125 – 141 .
- Terregino , C. , Cattoli , G. , Guberti , V. , De Nardi , R. , Scremin , M. , Beato , M.S. and Capua , I. 2005 . Isolation of influenza A viruses belonging to the H7N7 and H7N4 subtype from wild and domestic waterfowl in Italy . The Veterinary Record , 26 : 292
- Webster , R.G. and Hulse , D.J. 2004 . Microbial adaptation and change: avian influenza . Revue Scientifique et Technique , 23 ( 2 ) : 453 – 465 .