Abstract
Poult enteritis mortality syndrome (PEMS) of turkeys and runting-stunting syndrome (RSS) of chickens are significant viral enteric diseases of poultry. Although a number of different viruses, including avian reoviruses, rotaviruses, astroviruses and coronaviruses, have been isolated from the intestinal contents of birds in affected poultry flocks, their role in PEMS and RSS is not yet understood. Here, we report the application of a molecular screening method to detection of novel viruses in intestinal samples of chickens and turkeys exhibiting characteristic signs of enteric disease. The technique is based on random amplification of particle-associated nucleic acids in clinical samples. Using this method we successfully identified parvovirus DNA sequences in intestinal homogenates of affected birds. This is the first time partial genomic sequences of autonomously replicating chicken and turkey parvoviruses have been described. Sequence analysis of the left end of the genome, including the complete non-structural gene, demonstrated that the chicken and turkey parvoviruses were closely related to each other and were representative of a novel member of the Parvovirus family. These parvoviruses may play a significant role in the aetiology of PEMS and RSS.
Introduction
Viral enteric disease is a significant economic problem in the poultry industry throughout the world. The major enteric disease complexes in turkeys are known as poult enteritis complex (PEC) (Barnes et al., Citation2000) or the more severe poult enteritis mortality syndrome (PEMS), in which mortalities of up to 96% have been observed (Barnes & Guy, Citation2003), and in broiler chickens the complex is known as runting-stunting syndrome (RSS) (Page et al., Citation1982; Pass et al., Citation1982; Goodwin et al., Citation1993).
Enteric disease in poultry is characterized by diarrhoea, depression, ingestion of litter, increased vocalization and huddling. Morbidity and mortality are variable, and in many cases decreases in livability are due primarily to culling, but in some cases, such as with PEMS, there is substantial direct mortality (Barnes & Guy, Citation2003). During an outbreak, 80% to 90% of the flocks in an affected region may have clinical disease. The economic impact from enteric disease is primarily due to poor production, due to the failure of affected birds to grow, as well as due to the increase in costs of therapy, poor feed conversion efficiency and the increased time to reach target weights.
Improved management practices and antimicrobial therapy have been used, with limited success, to reduce the losses due to enteric disease (Barnes & Guy, Citation2003). However, there is no vaccine against the disease. Vaccine development is primarily hindered by the absence of a clearly defined aetiological agent (Barnes & Guy, Citation2003; Reynolds, Citation2003). A critical role for viruses in enteric disease is supported by the reproduction of the disease with preparations from the intestinal contents of affected birds that do not contain bacteria or protozoa (Barnes & Guy, Citation2003).
Viruses from numerous families, most frequently astroviruses, coronaviruses, reoviruses and rotaviruses, have been identified in the intestinal tracts of poultry with enteric disease (Reynolds et al., Citation1987; Guy, Citation1998; Koci et al., Citation2000; Spackman et al., Citation2005; Pantin-Jackwood et al., Citation2007a). Viruses from several other families, including enteroviruses and caliciviruses, have also been identified in intestinal samples of poultry with enteric disease (Theil & Saif, Citation1987; Guy & Barnes, Citation1991; Hayhow et al., Citation1993). However, viruses from each of these families have been identified in samples from flocks that appear healthy (Reynolds et al., Citation1987; Day et al., Citation2007; Pantin-Jackwood et al., Citation2007b), indicating that there could be pathotypic variation between these viruses. Currently, the role of these viruses in the aetiology of enteric disease of poultry is not understood.
In the mid-1980s, viral enteric disease of young turkeys characterized by stunting of affected birds, diarrhoea and increased mortality was described (Trampel et al., Citation1983). Electron microscopic examination of epithelial cells of the ileum revealed inclusion bodies containing hexagonal particles 15 to 20 nm in diameter that were tentatively identified as parvoviruses. Other studies also demonstrated the presence of parvovirus-like particles in intestinal homogenates of 10-day-old chickens with enteric disease (Kisary et al., Citation1984). Inoculation of 1-day-old broilers with purified viral particles resulted in characteristic clinical signs of RSS (Kisary, Citation1985). Subsequent molecular biological studies revealed that the virus belonged to the Parvovirus family (Kisary et al., Citation1985). Although parvoviruses are common pathogens in other species, the prevalence of parvoviruses in chicken and turkey flocks has not been confirmed using more definitive methods and the role of these viruses in the aetiology of enteric disease in poultry has not been further investigated.
In the present study we used a random hexamer polymerase chain reaction (PCR)-based approach to detect unknown viral sequences in enteric samples of chickens and turkeys with enteric disease.
Materials and Methods
Particle-associated nucleic acid amplification
To detect viruses of unknown sequence in clinical samples, we used a PCR approach allowing sequence-independent amplification (Allander et al., Citation2005; Stang et al., Citation2005). This random PCR resulted in representative amplification of all nucleic acid in the reaction. The specificity was not provided by the PCR but rather by selective isolation of particle-associated nucleic acids. The DNA from purified viral particles was extended using random primers containing a conserved 20-nucleotide 5′ sequence and a degenerate six-nucleotide 3′ end. Extended products were subsequently amplified by PCR with a primer complementary to only the conserved end of the random primer.
Enteric samples were collected in California, USA in 2004 from turkey poults with PEMS (Southeast Poultry Research Laboratory (SEPRL) sample collection, Athens, Georgia, USA). Forty-two intestinal homogenates (10% suspension in phosphate-buffered saline) were pooled and centrifuged at 775×g for 15 min at room temperature and the supernatants were collected. A homogenate of intestinal tissue from chickens inoculated with a parvovirus strain designated ABU virus was clarified by centrifugation at 775×g and the supernatant subjected to further processing. The ABU virus was isolated from chickens in Hungary in 1985 (Kisary et al., Citation1985), and was purified by caesium chloride density gradient centrifugation prior to inoculation of the chickens. Samples from chickens in the enteric sample collection of SEPRL collected in Georgia, USA in 2005 were used for PCR studies.
The supernatants were filtered through either a 0.45 µm (turkeys) or a 0.22 µm (chickens) syringe filter (Millipore) to remove bacterial cells. Filtered supernatants were treated with 10 U DNase I (Invitrogen) for 2 h at 37°C to remove unencapsidated DNA. Samples were then ultracentrifuged at 200 000×g for 90 min at 4°C on a 20% sucrose cushion. The resulting pellet was suspended in 500 µl TE buffer (10mM Tris-HCl and 1mM ethylenediamine tetraacetic acid, pH 8.0), and DNA was extracted from a 200 µl sample of the suspension using a total DNA Extraction kit (Qiagen). DNA was eluted in 200 µl elution buffer provided in the kit and the nucleic acid concentration was measured using a spectrophotometer (Nanodrop Technologies).
Extracted DNA (40 µl) was mixed with 5 µl of 10×Klenow buffer (New England Biolabs), 2 µl of a 10 µM solution of each dNTP and 2 µl of a 10 µM solution of primer FR26RV-N (GCCGGAGCTCTGCAGATATCNNNNNN). The reaction mix was incubated at 94°C for 4 min and on ice for 2 min, after which 2 U (1 µl) Klenow fragment of DNA polymerase (New England Biolabs) was added, and the reaction mix then incubated at 37°C for 1 h. This denaturation–annealing–elongation cycle was repeated once and then the Klenow fragment was inactivated at 75°C for 30 min. The DNA was purified with a PCR Purification kit (Qiagen) and eluted in 20 µl elution buffer (TE) provided by the manufacturer.
Five microlitres of the eluted DNA was used as a template in a subsequent PCR. For the turkey samples, the 50 µl reaction mix contained 1×Pfx reaction buffer (Invitrogen), 2.5 mM MgCl2, 0.2 mM each dNTP, 40 pmol primer FR20RV (GCCGGAGCTCTGCAGATATC) and 2 U Pfx DNA polymerase (Invitrogen). After 10 min incubation at 94°C, 30 cycles of amplification (94°C for 30 sec, 60°C for 1 min and 68°C for 3 min) were performed. The amplification product was purified using a Qiagen PCR Purification kit and eluted in 20 µl TE buffer. Five microlitres of the eluted, purified PCR product was ligated to the vector TOPO-Blunt (Invitrogen) and this was used to transform Escherichia coli TOP-10 cells (Invitrogen) according to the manufacturer's instructions. Bacteria were plated on Luria-Bertani (LB) agar plates containing kanamycin at 50 µg/ml.
For the ABU virus sample, HotStar Taq DNA polymerase was used (Qiagen) according to the manufacturer's protocol. The reaction mix contained 5 µl Klenow fragment amplified and purified ABU virus DNA, 25 µl HotStar mix and 40 pmol primer FR20RV. PCR and purification of the products were performed as described above. The purified PCR products were then separated in an agarose gel, and fragments between 500 and 2000 base pairs in length were excised and extracted using a Qiagen Gel Extraction kit (Qiagen). Five microlitres of the eluted DNA was ligated into the TOPO TA cloning vector (Invitrogen) and this was used to transform TOP-10 E. coli cells. Bacteria were then plated as described above.
Sequencing
Colonies were inoculated into LB broth (containing kanamycin) in 96-well plates and incubated at 37°C for 24 h in a shaker incubator. DNA was prepared using a PerfectPrep Plasmid purification kit (Eppendorf) and sequencing was performed with M13 forward and reverse primers, and the reaction products were separated on an AB-3730 automated DNA sequencer. The final DNA consensus sequence had an average eight-fold redundancy at each base position.
The sequences obtained were compared with those in the GenBank database using the BLASTn and BLASTx algorithms (http://www.ncbi.nlm.nih.gov/blast/Blast.cgi). For phylogenetic analysis, nucleotide and amino acid pairwise alignments were generated using Needleman–Wunsch global alignment (Labarga et al., Citation2007) and the ClustalW programs (Thompson et al., Citation1994). Phylogenetic and molecular evolutionary analyses were conducted using the MEGA version 4.0 software package (Tamura et al., Citation2007) (http://www.megasoftware.net/mega4). The GenBank accession numbers for the sequences determined in this study are EU304808 (chicken parvovirus) and EU304809 (turkey parvovirus).
PCR for chicken and turkey parvoviruses
DNA from individual enteric samples was extracted using a Qiagen DNA Extraction kit (Qiagen) in the presence of 5 µg carrier RNA. Extracted DNA (5 µl) was used as a template for PCR. The 50 µl reaction mix contained 25 µl HotStar Taq solution and 20 pmol of each of the following forward and reverse primers: NS1F, 5′-TATGATCTGTCAATGATTGACCAGCTGA-3′; NS2F, 5′-TCGGCTGCCAGTGCTGCTGAGCTGGTAA-3′; NS3F, 5′-CAACCAGTGAATCCATACTCATTTCTA-3′; NS1R, 5′-GTACCACTCGAGAGGGTTCAGGGAGAT-3′; and NS2R, 5′-CCAATAGGTCGATGGTTTCTGTAAGAT-3′. After 15 min incubation at 95°C, 30 cycles of amplification (94°C for 30 sec, 55°C for 1 min and 68°C for 3 min) were performed. PCR products were visualized after separation in an ethidium bromide stained agarose gel. The amplicons were purified and ligated into the TOPO TA cloning vector and this was used to transform TOP-10 E. coli cells. DNA preparation and sequencing were performed as described above.
Results
Detection of parvovirus by random PCR of particle-associated nucleic acid
A total of 384 transformant colonies were selected from the library obtained from particle-associated nucleic acid amplification from a pool of turkey intestinal homogenates and were subjected to sequence analysis. After removal of vector and poor-quality sequences, 146 (38%) clones remained for further studies. Of the 146 clones, a high number (n = 75) of sequences were found to contain primer dimers. The remaining 71 clones were categorized using nucleotide and translated BLAST searches into bacterial, phage, viral and unknown sequences. Fourteen (19%) clones had significant similarity to viral sequences. The sequences were similar to sequences from members of four different DNA virus families, the Adenoviridae, Herpesviridae, Poxviridae and Parvoviridae. PCR amplification using primers based upon the sequence of each clone was unsuccessful when used with DNA extracted from the original intestinal pool.
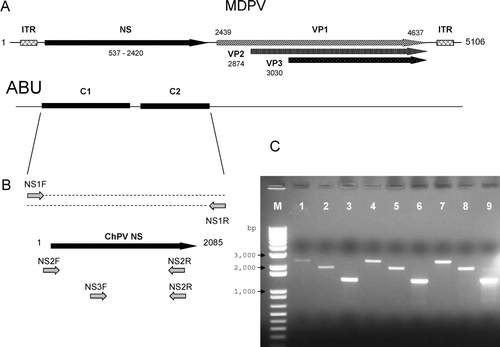
To improve the specificity and sensitivity of the technique, samples from chickens inoculated with the ABU virus were used. These birds showed characteristic signs of RSS at the time of sample collection. HotStar PCR polymerase was used to avoid primer dimer formation and only products between 500 and 2000 base pairs were cloned. A total of 384 clones from this library were sequenced. A high proportion of the clones (307/80%) were similar to chicken DNA, phages or viral sequences. Two contigs were assembled from 27 and 24 clones, respectively (a). Contig 1 was 1417 nucleotides and Contig 2 was 933 nucleotides. These sequences had no significant nucleotide similarity in the database, but the deduced amino acid sequences from both contigs were similar to those of members of the family Parvoviridae.
Figure 1. Detection of parvovirus DNA sequences in chicken and turkey enteric samples. 1a: Genome structure of Muscovy duck parvovirus (MDPV) showing the NS gene on the left and the capsid protein genes (VP1, VP2, VP3) on the right side, as well as the flanking inverted terminal repeats (ITR). Two contigs (C1 and C2) detected in intestinal homogenates of chickens inoculated with the ABU virus by random hexamer cloning are shown. 2b: PCR amplification of the ChPV NS gene using C1 and C2 specific primers. 1c: Agarose gel electrophoresis of PCR amplicons from ABU DNA (lanes 1, 2, 3), chicken enteric DNA (SEPRL collection) (lanes 4, 5, 6), and turkey enteric DNA (SEPRL collection) (lanes 7, 8, 9). The PCR primers were NS1F and NS1R (lanes 1, 4, 7), NS2F and NS2R (lanes 2, 5, 8), and NS3F and NS2R (lanes 3, 6, 9). M, 1 kb DNA marker (Invitrogen).
The non-structural gene of chicken and turkey parvoviruses
Both Contig 1 and Contig 2 sequences were successfully amplified from the original sample by PCR using the NS1F and NS1R primer sets (b). The size of the resultant amplicon was about 2.4 kb, indicating that the gap between the two contigs was less than 100 base pairs. The amplicon was cloned and sequenced to determine the full nucleotide sequence (c). The deduced amino acid sequence of this amplicon was similar to the left end of the genome of members of the Parvoviridae. Further analysis showed that the chicken parvovirus (ChPV) sequence (2407 nucleotides) contained the complete non-structural (NS) gene.
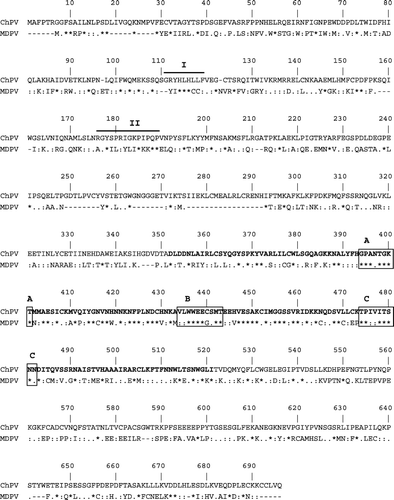
The ChPV NS protein had greatest similarity with the non-structural gene of the Muscovy duck parvovirus (); it was slightly longer (694 amino acids) than the duck NS protein (627 amino acids), and they had 133 (19.1%) identical, 152 (21.8%) highly similar and 93 (45.7%) weakly similar residues. The ChPV NS protein contained the replication initiator motif (I and II) (Ilyina & Koonin, Citation1992), helicase superfamily domains (A, B, and C) and nucleoside triphosphate (NTP)-binding motifs in the middle of the protein (Momoeda et al., Citation1994; Sukhumsirichart et al., Citation2006), which are highly conserved in the NS proteins of parvoviruses, as well as in various viral and cellular proteins (Iversen & Rhode III, Citation1990; Koonin, Citation1993).
Figure 2. Comparison of the amino acid sequence of the ChPV NS gene with the NS gene of Muscovy duck parvovirus (MDPV). Replication initiator signals I and II are shown with solid lines. The putative helicase domains (A, B, and C) and the NTP binding sites are boxed.
To screen for the presence of parvovirus sequences in turkey intestinal samples, PCRs were performed using the NSF and NSR primers, and the amplification products were visualized in agarose gels (c). All of the chicken NS sequence-based forward and reverse primers generated products from turkey enteric DNA samples and the size of each product were identical to that amplified from chicken enteric viral DNA with the same primers. Sequencing of the largest amplicon from the turkey samples (c) revealed that it contained the entire turkey parvovirus (TuPV) NS gene (2085 nucleotides) and that the gene had 98% identity at the nucleotide level with the chicken parvovirus NS gene. There were only three residues within the TuPV NS protein sequence that differed from the ChPV NS protein sequence. The parvovirus sequence was also detected in intestinal samples collected during enteric disease outbreaks in chickens in the USA using PCR amplification (c). Sequence analysis of these PCR products showed that the NS protein of these viruses had 100% amino acid sequence identity with the ABU chicken parvovirus NS protein sequence.
Phylogenetic analysis of chicken and turkey parvovirus NS genes
The chicken parvovirus NS gene sequence had significant similarity with the sequences of the NS genes of each of the previously described parvoviruses (46.0% to 54.3% similarity at the amino acid level and 42.9% to 47.4% identity at the nucleotide level).
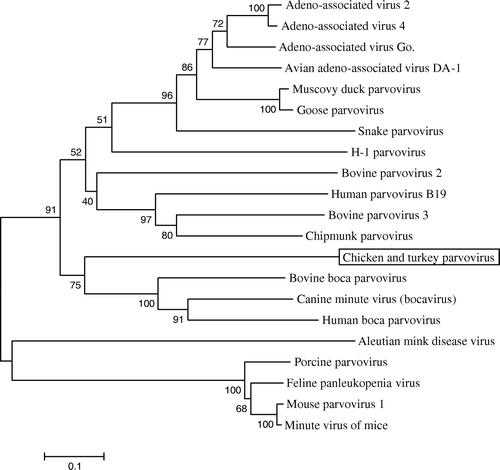
Because of the considerable sequence heterogeneity among the NS genes of these viruses, the most conserved region of the ChPV NS gene, a 170-amino-acid region located in the centre of the gene (corresponding to residues 352 to 521) (), was selected to construct a phylogenetic tree. Each of the different parvovirus genera formed a separate branch () (Lukashov & Goudsmit, Citation2001), with the chicken and turkey parvoviruses clustered with canine minute virus, bovine bocavirus 1 and human bocavirus. While these data suggest that ChPV and TuPV may be most closely related to members of the Bocavirus genus, the relatively short sequences (approximately 10% of the total genome) used for the analysis do not allow them to be definitively classified into any of the five recognized phylogenic groups.
Figure 3. Phylogenetic tree showing the relationship between the chicken and turkey parvoviruses and other members of the Parvovirinae subfamily. A conserved sequence of 170 residues of the chicken and turkey parvovirus NS gene was aligned with the sequences of other parvoviruses including the prototype viruses from each genera: minute virus of mice (genus Parvovirus), B19 human parvovirus (genus Erythrovirus), adeno-associated virus 2 (genus Dependovirus), Aleutian mink disease virus (genus Amdovirus), and bovine parvovirus 1 (genus Bocavirus). The evolutionary history was inferred using the neighbour-joining method (Saitou & Nei, Citation1987). The consensus tree from 1000 bootstrap replicates is shown. The percentage of trees that contained the consensus branch is also shown for each branch (Felsenstein, Citation1985). The scale at the bottom shows the number of substitutions inferred per site.
Discussion
In the present study, we detected parvovirus sequences in enteric samples of chickens with RSS and of turkeys with PEMS using a random hexamer PCR-based approach. Our sequence analysis indicated that these parvoviruses were closely related to each other and that they are representative of a novel member of the family Parvoviridae.
Electron microscopic studies have previously demonstrated the presence of parvovirus-like particles (ABU virus) in enteric samples of chickens with RSS (Kisary et al., Citation1984) and the ABU virus was classified as a novel parvovirus (Kisary et al., Citation1985), but efforts to isolate the virus in cell culture were unsuccessful (Kisary, unpublished data).
The ABU virus had previously been shown to be free of potential helper viruses, including adenoviruses and herpesviruses (Kisary et al., Citation1985), and the NS gene of the parvovirus did not have high levels of sequence similarity with the avian adeno-associated viruses that have been described previously. It is reasonable to consider this is the first report of a partial genomic sequence of an autonomously replicating chicken parvovirus.
Parvoviruses have a small linear single-stranded DNA genome between 4.5 and 5.5 kb in length encapsidated within a non-enveloped, icosahedral virion, approximately 20 nm in diameter. Classification of parvoviruses is now based on their genomic structure rather than the previous classical criteria, which included antigenicity and host restriction. Parvoviruses that infect vertebrate species have been classified into five genera: the parvoviruses, which include minute virus of mice as the prototype; the erythroviruses, of which the human parvovirus B19 is the prototype; the dependoviruses, which include the adeno-associated viruses and the goose and Muscovy duck parvoviruses; the amdoviruses, of which Aleutian mink disease virus is currently the only known member; and the bocaviruses, which include bovine parvovirus and canine minute virus (Tattersall et al., Citation2005).
The partial sequence (2407 nucleotides) of the chicken and turkey parvoviruses determined in this study comprised approximately 45% of the complete genome and included the entire NS gene. Amino acid sequence alignments revealed a significant similarity with several members of the parvoviruses, but there was not a particularly high level of similarity with any one of those viruses. These data indicated that the chicken and turkey parvoviruses are representative of an independent member of the Parvovirinae subfamily. When the complete NS protein sequences of the chicken and turkey parvoviruses were aligned with the NS protein of other parvoviruses, the similarity was closest to the Muscovy duck parvovirus, based on the total number of identical or similar residues (378 amino acids with the duck parvovirus and 348 amino acids with the bocaviruses). However, the phylogenetic tree was inferred using only the central conserved region of the NS protein (170 amino acids) and these data indicated a closer relationship with the bocaviruses. Further sequence analysis will be necessary to determine the exact relationship of ChPV and TuPV with each other and with other parvoviruses.
Allander et al. (Citation2001, 2005) discovered novel bovine and human parvoviruses within the Bocavirus genus of the Parvovirinae subfamily using the same technique as we have used. Their data suggested that reproducible detection of 105 virus copies was possible by sequencing 96 clones. In our studies, we sequenced 384 clones to assemble a 2.4 kb region of the chicken parvovirus genome. This suggests that the virus titre in the original sample of ABU virus was at least 104 particles/ml. It is possible that a lower virus titre might have contributed to the failure to assemble parvoviral contigs in the analyses of the pool of turkey enteric samples. Subsequent PCR studies suggested that this was probably the case. Primers specific for ChPV successfully amplified parvovirus sequences from turkey enteric samples, and this PCR product had a very high level of sequence identity with ChPV. The samples from chickens and turkeys used in this study were obtained from flocks that were completely isolated from each other geographically so this partial sequence analysis suggests recent divergence from a common ancestor, or possibly that the same viruses can infect both species. Further comparative sequencing data will be required to help determine the origin of these viruses and to establish whether they are distinct from each other.
The high levels of identity between the chicken and turkey parvoviruses appears greater than that seen between Muscovy duck and goose parvoviruses, the NS genes of which are 90% identical at the amino acid level. These parvoviruses have a high level of sequence identity (81.9%) across their entire genome (Zadori et al, Citation1995) and cause similar diseases in their respective hosts (Gough, Citation2003).
The role of chicken and turkey parvoviruses in disease is yet to be fully determined. It has been demonstrated that the ABU chicken parvovirus can induce clinical signs in broiler chickens characteristic of RSS (Kisary et al., Citation1985) but the pathogenicity of turkey parvovirus has not been investigated. We detected both viruses in intestinal samples from poultry with enteric disease, suggesting that the chicken and turkey parvoviruses may play a role in the aetiology of enteric diseases in poultry.
Acknowledgements
The authors thank Fenglan Li for her excellent technical assistance, and the sequencing unit at SEPRL for outstanding support.
References
- Allander , T. , Emerson , S.U. , Engle , R.E. , Purcell , R.H. and Bukh , J. 2001 . A virus discovery method incorporating DNase treatment and its application to the identification of two bovine parvovirus species . Proceedings of the National Academy of Sciences U S A , 98 : 11609 – 11614 .
- Allander , T. , Tammi , M.T. , Eriksson , M. , Bjerkner , A. , Tiveljung-Lindell , A. and Andersson , B. 2005 . Cloning of a human parvovirus by molecular screening of respiratory tract samples . Proceedings of the National Academy of Sciences U S A , 102 : 12891 – 12896 .
- Barnes , H.J. and Guy , J.S. 2003 . “ Poult enteritis-mortality syndrome ” . In Diseases of Poultry , 11th edn , Edited by: Saif , Y.M. , Barnes , H.J. , Glisson , J.R. , Fadly , A.M. , McDougald , L.R. and Swayne , D.E. 1171 – 1180 . Ames : Iowa State Press .
- Barnes , H.J. , Guy , J.S. and Vaillancourt , J.P. 2000 . Poult enteritis complex . Revue Scientifique et Technique , 19 : 565 – 588 .
- Day , J.M. , Pantin-Jackwood , M.J. and Spackman , E. 2007 . Sequence and phylogenetic analysis of the S1 genome segment of turkey-origin reoviruses . Virus Genes , 35 : 235 – 242 .
- Felsenstein , J. 1985 . Confidence limits on phylogenies: an approach using the bootstrap . Evolution , 39 : 783 – 791 .
- Goodwin , M.A. , Davis , J.F. , McNulty , M.S. , Brown , J. and Player , E.C. 1993 . Enteritis (so-called runting stunting syndrome) in Georgia broiler chicks . Avian Diseases , 37 : 451 – 458 .
- Gough , R.E. 2003 . “ Goose parvovirus infection ” . In Diseases of Poultry , 11th edn , Edited by: Saif , Y.M. , Barnes , H.J. , Glisson , J.R. , Fadly , A.M. , McDougald , L.R. and Swayne , D.E. 367 – 374 . Ames : Iowa State Press .
- Guy , J.S. 1998 . Virus infections of the gastrointestinal tract of poultry . Poultry Science , 77 : 1166 – 1175 .
- Guy , J.S. and Barnes , H.J. 1991 . Partial characterization of a turkey enterovirus-like virus . Avian Diseases , 35 : 197 – 203 .
- Hayhow , C.S. , Saif , Y.M. , Kerr , K.M. and Whitmoyer , R.E. 1993 . Further observations on enterovirus infection in specific-pathogen-free turkey poults . Avian Diseases , 37 : 124 – 134 .
- Ilyina , T.V. and Koonin , E.V. 1992 . Conserved sequence motifs in the initiator proteins for rolling circle DNA replication encoded by diverse replicons from eubacteria, eucaryotes and archaebacteria . Nucleic Acids Research , 20 : 3279 – 3285 .
- Iversen , P. and Rhode III , S.L. 1990 . “ Parvovirus proteins ” . In Handbook of Parvoviruses , Edited by: Tijssen , P. 81 – 90 . Boca Raton , FL : CRC Press .
- Kisary , J. 1985 . Experimental infection of chicken embryos and day-old chickens with parvovirus of chicken origin . Avian Pathology , 14 : 1 – 7 .
- Kisary , J. , Nagy , B. and Bitay , Z. 1984 . Presence of parvoviruses in the intestine of chickens showing stunting syndrome . Avian Pathology , 13 : 339 – 343 .
- Kisary , J. , Avalosse , B. , Miller-Faures , A. and Rommelaere , J. 1985 . The genome structure of a new chicken virus identifies it as a parvovirus . Journal of General Virology , 66 : 2259 – 2263 .
- Koci , M.D. , Seal , B.S. and Schultz-Cherry , S. 2000 . Molecular characterization of an avian astrovirus . Journal of Virology , 74 : 6173 – 6177 .
- Koonin , E.V. 1993 . A common set of conserved motifs in a vast variety of putative nucleic acid-dependent ATPases including MCM proteins involved in the initiation of eukaryotic DNA replication . Nucleic Acids Research , 21 : 2541 – 2547 .
- Labarga , A. , Valentin , F. , Andersson , M. and Lopez , R. 2007 . Web Services at the European Bioinformatics Institute . Nucleic Acids Research , 35 : 1 – 6 .
- Lukashov , V.V. and Goudsmit , J. 2001 . Evolutionary relationships among parvoviruses: virus–host coevolution among autonomous primate parvoviruses and links between adeno-associated and avian parvoviruses . Journal of Virology , 75 : 2729 – 2740 .
- Momoeda , M. , Wong , S. , Kawase , M. , Young , N.S. and Kajigaya , S. 1994 . A putative nucleoside triphosphate-binding domain in the nonstructural protein of B19 parvovirus is required for cytotoxicity . Journal of Virology , 68 : 8443 – 8446 .
- Page , R.K. , Fletcher , O.J. , Rowland , G.N. , Gaudry , D. and Villegas , P. 1982 . Malabsorption syndrome in broiler chickens . Avian Diseases , 26 : 618 – 624 .
- Pantin-Jackwood , M.J. , Spackman , E. and Day , J.M. 2007a . Pathology and virus tissue distribution of turkey origin reoviruses in experimentally infected turkey poults . Veterinary Pathology , 44 : 185 – 195 .
- Pantin-Jackwood , M.J. , Spackman , E. , Day , J.M. and Rives , D. 2007b . Periodic monitoring of commercial turkeys for enteric viruses indicates continuous presence of astrovirus and rotavirus on the farms . Avian Diseases , 51 : 674 – 680 .
- Pass , D.A. , Robertson , M.D. and Wilcox , G.E. 1982 . Runting syndrome in broiler chickens in Australia . Veterinary Record , 110 : 386 – 387 .
- Reynolds , D.L. 2003 . “ Multicausal enteric diseases ” . In Diseases of Poultry , 11th edn , Edited by: Saif , Y.M. , Barnes , H.J. , Glisson , J.R. , Fadly , A.M. , McDougald , L.R. and Swayne , D.E. 1169 – 1170 . Ames : Iowa State Press .
- Reynolds , D.L. , Saif , Y.M. and Theil , K.W. 1987 . Enteric viral infections of turkey poults: incidence of infection . Avian Diseases , 31 : 272 – 276 .
- Saitou , N. and Nei , M. 1987 . The neighbor-joining method: a new approach for reconstructing phylogenetic trees . Molecular Biology and Evolution , 4 : 406 – 425 .
- Spackman , E. , Kapczynski , D. and Sellers , H. 2005 . Multiplex real-time reverse transcription-polymerase chain reaction for the detection of three viruses associated with poult enteritis complex: turkey astrovirus, turkey coronavirus, and turkey reovirus . Avian Diseases , 49 : 86 – 91 .
- Stang , A. , Korn , K. , Wildner , O. and Uberla , K. 2005 . Characterization of virus isolates by particle-associated nucleic acid PCR . Journal of Clinical Microbiology , 43 : 716 – 720 .
- Sukhumsirichart , W. , Attasart , P. , Boonsaeng , V. and Panyim , S. 2006 . Complete nucleotide sequence and genomic organization of hepatopancreatic parvovirus (HPV) of Penaeus monodon . Virology , 346 : 266 – 277 .
- Tamura , K. , Dudley , J. , Nei , M. and Kumar , S. 2007 . MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0 . Molecular Biology and Evolution , 24 : 1596 – 1599 .
- Tattersall , P. , Bergoin , M. , Bloom , M.E. , Brown , K.E. , Linden , R.M. , Muzyczka , N. , Parrish , C.R. and Tijssen , P. 2005 . “ Taxonomic structure of the family Parvoviridae ” . In Eighth Report of the International Committee on Taxonomy of Viruses , Edited by: Fauquet , C.M. , Mayo , M.A. , Maniloff , J. , Desselberger , U. and Ball , L.A. 353 – 369 . San Diego , CA : Elsevier .
- Theil , K.W. and Saif , Y.M. 1987 . Age-related infections with rotavirus, rotaviruslike virus, and atypical rotavirus in turkey flocks . Journal of Clinical Microbiology , 25 : 333 – 337 .
- Thompson , J.D. , Higgins , D.G. and Gibson , T.J. 1994 . CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice . Nucleic Acids Research , 22 : 4673 – 4680 .
- Trampel , D.W. , Kinden , D.A. , Solorzano , R.F. and Stogsdill , P.L. 1983 . Parvovirus-like enteropathy in Missouri turkeys . Avian Diseases , 27 : 49 – 54 .
- Zadori , Z. , Stefancsik , R. , Rauch , T. and Kisary , J. 1995 . Analysis of the complete nucleotide sequences of goose and muscovy duck parvoviruses indicates common ancestral origin with adeno-associated virus 2 . Virology , 212 : 562 – 573 .