Abstract
A TaqMan®-based real-time, quantitative polymerase chain reaction (qPCR) assay utilizing the mgc2 gene was developed to detect Mycoplasma gallisepticum in conjunctival swabs of experimentally infected house finches. The assay was demonstrated to be quantitative by the standard curve method with reproducible results within runs and between runs. The detection limit of the mgc2 assay was examined using two standards. The test had a detection limit of less than 14 copies per reaction when tested with a plasmid standard and less than 10 copies per reaction when tested with M. gallisepticum genomic DNA. All M. gallisepticum-negative birds (10 specific pathogen free chickens and 10 house finches) were negative by mgc2 qPCR assay. Existing evidence suggests that an important part of M. gallisepticum pathogenesis includes both its attachment to and invasion of host cells. Thus, our test also made use of rag-1 as an internal control gene. The rag-1 qPCR results showed that host cell quantity varied greatly between conjunctival samples. After inoculation, M. gallisepticum levels in the house finch conjunctiva increased over the 7-day period post infection. The bird with the most pronounced clinical conjunctivitis harboured the highest level of M. gallisepticum and the bird that did not develop conjunctivitis had very low numbers of M. gallisepticum. Thus, it appears that development of conjunctivitis may correlate with M. gallisepticum load.
Introduction
Mycoplasma gallisepticum is a prokaryotic pathogen that causes chronic respiratory disease in chickens and infectious sinusitis in turkeys. Disease resulting from M. gallisepticum infection has considerable economic consequences for the poultry industry worldwide. In 1994, M. gallisepticum was detected in house finches (Carpodacus mexicanus) in the eastern part of North America (Ley et al., Citation1996). The infection caused major declines throughout the eastern population and eventually spread to the native western range of the house finch (Ley et al., Citation2006). The most prominent clinical sign of infection in finches is unilateral or bilateral conjunctivitis, which in experimentally infected birds develops approximately 4 days post infection (p.i.) (Kollias et al., Citation2004).
Several real-time quantitative polymerase chain reaction (qPCR) assays have been developed to measure M. gallisepticum in tracheal and palatine cleft swabs of domestic chickens. Two SYBR Green-based assays that use standard curves based on colony-forming units (CFUs) have been described. One assay utilizes the 16S rRNA gene (Mekkes & Feberwee, Citation2005), and the other utilizes the mga_0319 lipoprotein gene (Carli & Eyigor, Citation2003). Problems associated with these assays have been a lack of sensitivity or cross-reaction with other Mycoplasma species. Callison et al. (Citation2006) developed a TaqMan®-based assay that quantified M. gallisepticum based on mga_0319 gene copy numbers using a plasmid standard, instead of CFUs.
Lacking in the previously developed qPCR assays is the use of a host housekeeping gene. The measurement of a host gene is an important control to account for differences in swabbing methods, DNA extraction efficiencies, and inflammatory responses. There is also increasing evidence for the close association of M. gallisepticum with host cells, including both adhesion to and invasion of host cells. In other Mycoplasma species, adhesion to host cells plays a vital role in pathogenesis (Balish & Krause, Citation2005). This is also true for M. gallisepticum, as genes and corresponding proteins involved in cytadhesion have been identified and characterized (Keeler et al., Citation1996; Goh et al., Citation1998; Hnatow et al., Citation1998; Papazisi et al., Citation2002). There is also evidence that M. gallisepticum has the ability to invade host cells. Winner et al. (Citation2000) demonstrated that M. gallisepticum is internalized by human epithelial cells and chicken embryo fibroblasts in vitro. More recently, studies by Vogl et al. (Citation2008) provided evidence that M. gallisepticum can invade and adhere to chicken erythrocytes in vitro and can invade erythrocytes in vivo after aerosol inoculation.
In many polymerase chain reaction (PCR) assays, the 18S rRNA gene has been used as an endogenous control for eukaryotic DNA (for example, Bai et al., Citation2004; Casabianca et al., Citation2004) and RNA (Li et al., Citation2005). However, the use of this gene as a DNA control has several disadvantages. The gene is present in multiple copies within the eukaryotic genome and copy numbers vary among species and at the individual level (Long & Dawid, Citation1980). For example, the 18S rRNA copy number varies between chicken strains and among individuals within a given strain. Delany (Citation2000) found copy numbers ranging from 302 to 422 in broiler lines. Moreover, the 18S rRNA copy number in the house finch is unknown; therefore, quantification of host cell number is not feasible using this gene.
Conventional PCR has been used to identify infected house finches using conjunctival swabs. The mgc2 gene, which encodes a cytadhesin protein (Hnatow et al., Citation1998), is currently the one of the preferred gene targets for this assay, due to its specificity for M. gallisepticum (Garcia et al., Citation2005). We selected this gene for the development of our qPCR to quantify the level of M. gallisepticum infection in house finches. The recombination-activating gene 1 (rag-1), which is present in two copies/diploid cell in the genome of higher vertebrates (Groth & Barrowclough, Citation1999), was selected to quantify the number of host cells in conjunctival swab samples.
Materials and Methods
Sample sources
Serum samples and conjunctival samples from both eyes were obtained from 10 chickens from the specific pathogen free P2a (Weinstock & Schat, Citation1987) flock at Cornell University. Conjunctival samples were collected using cotton-tipped wooden-handle swabs (product #14-960-3N; Fisher Scientific, Pittsburgh, Pennsylvania, USA), placed in 300 µl tryptose phosphate broth and kept at 4°C until the swab was discarded. Then the remaining sample was stored at −28°C until DNA extraction. Blood was collected from the wing vein on the date of swab collection and also 4 weeks later to confirm M. gallisepticum-negative antibody status using serum plate agglutination assays (Charles River Laboratories, Wilmington, Massachusetts, USA). The P2a line has been negative for M. gallisepticum antibodies for at least 20 years and is tested monthly to confirm its negative status.
One-year-hatchling house finches were trapped in Tompkins County, New York, USA (42°51′N, 76°34′W) during October and November 2006 using mist nets under permits (LCP 99-039) from the New York State Department of Environmental Conservation (Albany, New York, USA) and a federal collecting permit (PRT 802829). Birds were placed in quarantine for a minimum of 2 weeks, and blood was collected from the wing vein into lithium-heparinized microcapillary tubes and screened for the presence of M. gallisepticum-specific antibody using the serum plate agglutination test (Intervet, Millsboro, Delaware, USA) (Kleven, Citation1998). The house finches were kept in individual wire bar cages with metal barriers between them to prevent mechanical spread of M. gallisepticum. Water and a pelleted diet (Roudybush, Inc., Cameron Park, California, USA) were provided ad libitum.
Ten additional 1-year-hatchling house finches were sampled during October 2007. After M. gallisepticum antibody-negative status was confirmed with serum plate agglutination, conjunctival swabs were obtained to serve as M. gallisepticum-negative samples. Samples from the left and right eyes were pooled for each bird.
Experimental infection
Five birds were inoculated in the conjunctival sac of the right and left eyes with 50 µl M. gallisepticum inoculum (3.24 x 105 CFU/ml) at day 0. Conjunctival swabs were collected at day 0 (pre-inoculation), and 1, 3, 5, and 7 days p.i. Conjunctival samples were collected using cotton-tipped swabs, placed in 300 µl tryptose phosphate broth, and kept at 4°C until the swab was removed. The remaining sample was then stored at −28°C until DNA extraction. Samples for the right and left eyes were processed separately so that the M. gallisepticum load for each eye could be determined. Physical signs of M. gallisepticum infection were quantified by scoring the severity of the inflammatory process in each eye using a 0 to 3 scale as described by Sydenstricker et al. (Citation2006): 0 = no visible inflammation; 1 = pink conjunctival discolouration and slight periorbital oedema, 2 = pink conjunctival discolouration, slight to moderate periorbital oedema, and epiphora with mucoid discharge; and 3 = red conjunctiva, epiphora and feather matting, feather loss around periorbital ring, severe conjunctival oedema, and at least some chemosis or rhinitis. Both eyes were scored on days 0, 1, 3, 5, and 7.
M. gallisepticum isolate
The M. gallisepticum inoculum used in the experiment was the sixth in vitro broth (Frey's medium with 15% swine serum, modified from Kleven [Citation1998]) passage from the original M. gallisepticum house finch isolate, ADRL 7994-1 (Ley et al., Citation1996). The viable count of this inoculum (ADRL 7994-1 6P) was 3.24 x 105 CFU/ml as determined by colony counts on agar from serial dilutions (Kollias et al., Citation2004). The M. gallisepticum utilized for assay development and sequencing was the seventh in vitro broth passage from M. gallisepticum house finch isolate, ADRL 7994-1.
DNA extraction and sample preparation
DNA extraction from conjunctival swab samples was carried out using a Qiagen DNeasy blood and tissue kit (Qiagen, Valencia, California, USA), following the manufacturer's recommended protocol for the purification of total DNA from animal tissues. The samples of experimentally infected house finches were diluted to a DNA concentration of 50 ng/5 µl for use in the qPCR. The DNA concentration was determined using a Bio-Rad SmartSpec™ 3000 (Bio-Rad, Hercules, California, USA) following the manufacturer's protocol. The samples of M. gallisepticum-negative birds were not further diluted because spectrophotometer readings of these samples were very low and accurate quantification of DNA in the samples was not feasible.
Preparation of DNA standards
A 303 base pair (bp) region of the mgc2 gene was amplified from the genome of M. gallisepticum strain ADRL 7994.1 by PCR with previously published primer sequences (Garcia et al., Citation2005) using the following PCR parameters: 94°C for 3 min; followed by 35 cycles of 94°C for 30 sec; 58°C for 30 sec, 72°C for 60 sec; followed by one cycle of 72°C for 10 min. A 1100 bp region of rag-1 was amplified from house finch, American goldfinch (Carduelis tristis), house sparrow (Passer domesticus), and chicken (Gallus gallus) DNA with the R17 and R22 primers published by Groth & Barrowclough (Citation1999). The PCR amplification parameters were: 5 min at 94°C; followed by 35 cycles of 94°C for 30 sec; 55°C for 40 sec, 72°C for 60 sec; followed by one cycle of 72°C for 10 min. A 119 bp fragment of the house sparrow 18S rRNA gene was amplified with conserved primer sequences (Lopez-Andreo et al., Citation2005) using the same PCR parameters as described for rag-1. The PCR products were cloned in the pCR®4-TOPO vector (Invitrogen, Carlsbad, California, USA) following the manufacturer's protocol. Random colonies were grown overnight in Luria-Bertani broth containing ampicillin, and plasmid DNA was extracted using a Qiagen MiniPrep kit (Qiagen) following the standard kit protocol. The insert was then sequenced (Automated 3730 DNA Analyzer; Applied Biosystems, Foster City, California, USA) and purified plasmid DNA from one clone containing the correct insert was used to construct the standard curves. For the rag-1 assay standard, plasmid was harvested from a clone containing the house finch rag-1 insert. The plasmid standard for the nitric oxide synthase 2 (nos2) assay was kindly provided by K.W. Jarosinski, Department of Microbiology and Immunology, Cornell University (Jarosinski et al., Citation2002). M. gallisepticum genomic DNA was extracted from the inoculum (ADRL 7994.1) utilized in this study to serve as an additional standard for the mgc2 qPCR. Fresh 10-fold dilutions were made from standard stocks in DNase-free water.
Primer and probe design
A 303 bp sequence of the mgc2 gene of isolate ADRL 7994-1 was aligned with mgc2 sequences of M. gallisepticum isolated from six house finches and one goldfinch from various locations and years. Primers and probes were designed with Primer Express (version 2.0; Applied Biosystems). Primers and probes for the real-time assays were synthesized by Integrated DNA Technologies (Coralville, Iowa, USA). Probes had 6-FAM on the 5′ end and BHQ-1 on the 3′ end—with the exception of the nos2 probe, which had TET on the 5′ end and TAMRA on the 3′ end.
Lasergene sequence analysis software (DNAstar, Madison, Wisconsin, USA) was utilized to create an alignment of rag-1 sequences of two house finches, a goldfinch, a house sparrow and a chicken. Primers and probe were chosen to be in areas of 100% identity in the house finch and goldfinch to ensure primers anneal to areas conserved among house finches. presents a list of the primer and probe sequences used in this study.
Table 1. Real-time primer and probe sequences utilized in this study
Real-time assays
The 20 µl reaction was performed in an ABI Prism 7500 (Applied Biosystems). The “fast” reaction utilized 10 µl TaqMan® Fast Universal PCR Master Mix (2x), No AmpErase® UNG (Applied Biosystems), 0.18 µl each of 100µM forward and reverse primers, 0.5 µl of 10 µM probe, 4.14 µl DNase-free water, and 5 µl sample. Samples were run in triplicate. Cycling parameters were 95°C for 20 sec and 40 cycles of 95°C for 3 sec and 60°C for 30 sec. The automatic threshold settings were used in analysis of samples. Samples were positive for M. gallisepticum if the cycle threshold (Ct) value was less than 40.
Nucleotide sequence accession numbers
Sequences can be accessed from GenBank (http://www.ncbi.nlm.nih.gov) with the following accession numbers: M. gallisepticum strain 7994-1 mgc2 (EF462343), house sparrow 18S rRNA gene (EF462342), and rag-1 sequences from two house finches (EU165349, EU165350), a chicken (EU165351), a house sparrow (EU165352), and a goldfinch (EU165353). The mgc2 sequences used in alignment of isolates from free-living finches included GenBank accession numbers AY556264, AY556259, AY556257, AY556245, AY556242, AY556240, and AY556233.
Statistical analysis
JMP (version 7.0; SAS Institute) was utilized for the least squares regression analysis presented later in . We report coefficient estimates (β), P values, and R 2 values.
Results
Primers and probe
The ADRL 7994-1 partial mgc2 sequence was found to be identical to that of the seven other house finch and goldfinch isolate sequences available in GenBank, and the real-time primers and probe () were developed to amplify a 227 bp region within the 303 bp region. The approximate size was confirmed by running real-time products on an agarose gel (data not shown). A BLAST search with the mgc2 real-time primers revealed significant alignments with a multitude of M. gallisepticum isolates, and no alignments with other Mycoplasma species. When the mgc2 sequence of ADRL 7994-1 and several M. gallisepticum reference strains were compared, several nucleotide polymorphisms were present in the qPCR target region (data not shown).
Standard curves
The mgc2 standard was based on 10-fold serial dilutions of plasmid containing a 303 bp mgc2 insert. The curve was created using 3.41 x 101 to 3.41 x 108 copy numbers. Serial 10-fold dilutions of the M. gallisepticum genomic DNA were used to create the curve using 1.00 x 100 to 1.00 x 106 copy numbers. The rag-1 and 18S rRNA gene standards were based on 10-fold serial dilutions of plasmids containing the 1100 bp house finch rag-1 and the 119 bp house sparrow 18S rRNA gene inserts, respectively. To generate the standard curves for rag-1 and 18S rRNA, 3.76 x 101 to 3.76 x 107 and 1.40 x 101 to 1.40 x 107 copy numbers, respectively, were used. Standard curves were generated for each run. The following standard curve equations represent typical runs: mgc2 plasmid, y= − 3.382x+39.98, R 2=0.998; M. gallisepticum genomic, y= − 3.723x+39.885, R 2=0.996; rag-1, y= − 3.504x+39.16, R 2=0.997; and 18S rRNA gene, y= − 3.578x+46.741, R 2=0.987 .
Assay detection limit and reproducibility
The mgc2 assay detection limit was determined using two standards, a plasmid standard and M. gallisepticum genomic DNA. The assay exhibited a detection limit of less than 14 copies per reaction using the plasmid standard and less than 10 copies per reaction using the genomic DNA standard. The inter-assay reproducibility of the mgc2 plasmid standard and genomic DNA standard () was determined using the mean Ct values from three independent runs, conducted on different days with new standard dilutions for each run. The intra-assay reproducibility of the mgc2 standard curve () is based on the mean Ct values from the standard curve run in triplicate. The linearity of the mgc2 and M. gallisepticum genomic DNA standard curves is shown in , with each curve representing the mean inter-assay Ct values presented in . The data demonstrate that the mgc2 assay is highly reproducible.
Figure 1. Linearity of standard curve. 1a: Standard curve based on mgc2 plasmid. 1b: Standard curve based on M. gallisepticum genomic DNA.
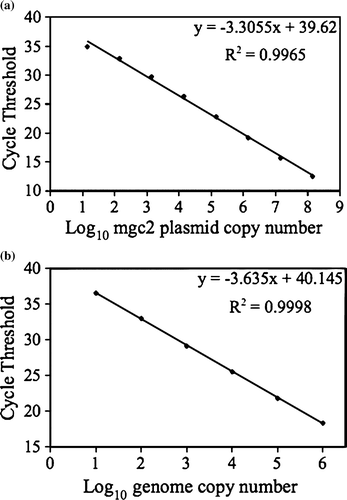
Table 2. Reproducibility and limit of detection of the standard curves used for the mgc2 assay
Control samples
Sensitive tests for the presence of M. gallisepticum-specific antibodies in passerine birds are lacking, and therefore it is not possible to obtain absolute confirmation that free-living passerine birds are truly negative for M. gallisepticum. Thus, we first confirmed that the mgc2 assay tested negative with avian DNA by using M. gallisepticum-negative chicken swab samples. The 10 chickens and 10 house finches, all free of M. gallisepticum infection based on the absence of antibodies, were also negative using mgc2 real-time PCR with Ct values >40. Nos2 was used as a host DNA control for the chicken swab samples to ensure DNA was efficiently extracted. The Nos2 copy number ranged from 363 to 40 000 copies/5 µl, with a mean copy number of 6509 and a standard deviation of 9177 copies. Rag-1 was used as the house finch DNA control. The Rag-1 copy number ranged from 13 to 1520 copies/5 µl, with a mean copy number of 255 and a standard deviation of 470 copies.
Quantification of M. gallisepticum load in conjunctival samples of five infected house finches
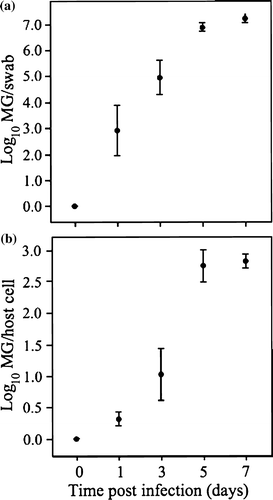
After experimental infection with M. gallisepticum, Bird 5 showed clinical signs by day 3, and Birds 2, 3, and 4 showed signs by day 7. Bird 1 did not develop clinical signs. a illustrates the mean log10 of total M. gallisepticum eluted from the swab as a function of time p.i. in birds that developed clinical signs (Birds 2, 3, 4, 5). Total M. gallisepticum eluted from the swab increases with time p.i. (β = 0.985, P<0.0001, R 2=0.80). In b, the mean log10 of M. gallisepticum copy number per house finch cell is plotted as a function of time p.i. in birds that developed clinical signs. M. gallisepticum/host cell increases with time p.i. (β = 0.450, P<0.0001, R 2=0.84). For each bird, the mean value of the right and left eyes was used to construct the plots in .
Figure 2. 2a: Mean M. gallisepticum (MG) load present in total swab samples of the conjunctiva of four inoculated house finches that developed clinical conjunctivitis, as a function of time p.i., with standard error bars. 2b: Mean M. gallisepticum (MG) load shown as the log10 value of mgc2 copy number per host cell, with standard error bars.
A summary of maximal M. gallisepticum load and eye score at day 7 post-infection is also presented in , showing data without log transformation. shows individual data for each bird, with the M. gallisepticum load and eye score plotted over the 7 days p.i. Bird 5 exhibited the earliest eye score at 3 days p.i. and also had the highest M. gallisepticum load. Although there is variation among individuals, these data show that the right and left eyes of individual birds experience similar M. gallisepticum load. also illustrates that while birds had similar M. gallisepticum loads throughout the 7-day period (e.g. Birds 2 and 5), the clinical signs developed at different time points. There are three data points excluded from the analysis. Samples from Bird 1 (day 3, right eye) and Bird 4 (day 7, right eye) could not be analysed because of insufficient amount of DNA in the sample. It was also suspected that the sample of Bird 1 at day 0 (left eye) was contaminated during the DNA extraction process, and this data point was not included in the graph.
Figure 3. Data from individual birds showing M. gallisepticum (MG) load over time and occurrence of conjunctivitis. Solid circles, an eye score of 1 on the day of conjunctival sampling; dashed circle, a more severe eye score of 2. R, right eye; L, left eye. Eye score code: 0 = no visible inflammation; 1 = pink conjunctival discolouration and slight periorbital oedema, 2 = pink conjunctival discolouration, slight to moderate periorbital oedema, and epiphora with mucoid discharge.
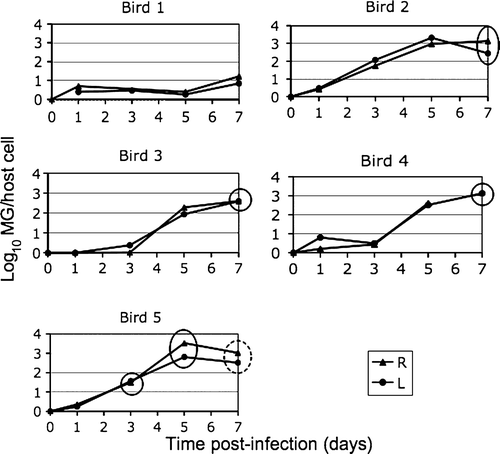
Table 3. Summary of maximal mycoplasmal load and eye score at day 7 p.i.
Host control genes
A BLAST search of the house sparrow 18S rRNA gene sequence confirmed that it had 100% sequence identity to many other avian species, including chicken. Thus, it was an appropriate standard in the house finch 18S rRNA gene qPCR assay. Among the house finches infected with M. gallisepticum, the average 18S rRNA gene copy number ranged from 1.57 x 102 to 1.40 x 108 (mean = 2.40 x 106±1.82 x 107) in 50 ng DNA. House finch rag-1 copy numbers were used to standardize the M. gallisepticum conjunctival load. Since rag-1 is present in two copies per diploid cell, the number of host cells in each sample was calculated by dividing the rag-1 copy number by two. In samples from experimentally infected birds (50 ng DNA/assay), the house finch cell number ranged from 7 to 2893 (mean = 212±487).
Discussion
We have developed a TaqMan®-based assay to detect M. gallisepticum in conjunctival swab samples of experimentally infected house finches. The standard curve was reproducible both between runs and within runs. The detection limit of the mgc2 assay was examined using two standards: M. gallisepticum genomic DNA extracted from culture, and a plasmid-based standard consisting of a partial mgc2 gene inserted into a bacterial vector. The test has a detection limit of less than 14 copies per reaction when tested with the plasmid standard and less than 10 copies per reaction when tested with the genomic DNA standard. All M. gallisepticum-negative birds (10 chickens and 10 house finches) resulted in a negative mgc2 assay result.
Our test also includes an internal control gene. The occurrence of conjunctivitis in infected finches makes the use of a host gene even more imperative, as host cells are being actively recruited to site of infection and may influence absolute quantity of the sample obtained. The evidence that M. gallisepticum adheres to host cells and has the ability to invade cells further underscores the importance measuring the number of host cells collected in the conjunctival swab samples. Because there are potentially hundreds of 18S rRNA gene copies in the house finch genome, we decided to explore single-copy gene alternatives and found rag-1 to be an acceptable host control gene that allows for quantification of host cells in the conjunctival samples. Our rag-1 results demonstrate that host cell quantity varies quite substantially between conjunctival samples. The use of rag-1 ensures that variation in samples due to swabbing method, infection status (e.g. birds with and without conjunctivitis), and any differences in DNA extraction methods are taken into account.
After experimental inoculation, M. gallisepticum levels in the house finch conjunctiva increase exponentially over the 7-day period p.i. The bird with the most severe conjunctivitis experienced the highest level of M. gallisepticum and the bird that did not develop conjunctivitis had a very low level of M. gallisepticum. Thus, it appears that development of conjunctivitis may correlate with M. gallisepticum load.
In summary, our real-time assay can provide sensitive detection of M. gallisepticum in conjunctival samples of house finches. The assay allows for the quantification of M. gallisepticum genome copy number while controlling for intrinsic differences that exist as a result of variations in sampling method and host inflammatory response. We hope this assay will serve as a valuable tool for future studies of M. gallisepticum pathogenesis and transmission in house finches.
Acknowledgements
The present study was funded through NSF grant EF-06227054 (A. Dhondt ) under the NSF-NIH Ecology of Infectious Diseases Program. The M. gallisepticum inoculum used in this study was provided by Dr David Ley, Department of Population Health and Pathobiology, College of Veterinary Medicine, North Carolina State University, Raleigh, North Carolina, USA.
References
- Bai , R.K. , Perng , C.L. , Hsu , C.H. and Wong , L.J.C. 2004 . Quantitative PCR analysis of mitochondrial DNA content in patients with mitochondrial disease . Annals New York Academy of Sciences , 1011 : 304 – 309 .
- Balish , M.F. and Krause , D.C. 2005 . “ Mycoplasma attachment organelle and cell division ” . In Mycoplasmas: Molecular Biology, Pathogenicity and Strategies for Control , Edited by: Blanchard , A. and Browning , G. 189 – 237 . Wymondham, Norfolk , , UK : Horizon Biosciences .
- Callison , S.A. , Riblet , S.M. , Sun , S. , Ikuta , N. , Hilt , D. , Leiting , V. , Kleven , S.H. , Suarez , D.L. and Garcia , M. 2006 . Development and validation of a real-time Taqman polymerase chain reaction assay for the detection of Mycoplasma gallisepticum in naturally infected birds . Avian Diseases , 50 : 537 – 544 .
- Carli , K.T. and Eyigor , A. 2003 . Real-time polymerase chain reaction for detection of Mycoplasma gallisepticum in chicken trachea . Avian Diseases , 47 : 712 – 717 .
- Casabianca , A. , Orlandi , C. , Fraternale , A. and Magnani , M. 2004 . Development of a real-time PCR assay using SYBR Green I for provirus load quantification in a murine model of AIDS . Journal of Clinical Microbiology , 42 : 4361 – 4364 .
- Delany , M.E. 2000 . Patterns of ribosomal gene variation in elite commercial chicken pure line populations . Animal Genetics , 31 : 110 – 116 .
- Garcia , M. , Ikuta , N. , Levisohn , S. and Kleven , S.H. 2005 . Evaluation and comparison of various PCR methods for detection of Mycoplasma gallisepticum infection in chickens . Avian Diseases , 49 : 125 – 132 .
- Goh , M.S. , Gorton , T.S. , Forsyth , M.H. , Troy , K.E. and Geary , S.J. 1998 . Molecular and biochemical analysis of a 105 kDa Mycoplasma gallisepticum cytadhesin (GapA) . Microbiology , 144 : 2971 – 2978 .
- Groth , J.G. and Barrowclough , G.F. 1999 . Basal divergences in birds and the phylogenetic utility of the nuclear RAG-1 gene . Molecular Phylogenetics and Evolution , 12 : 115 – 123 .
- Hnatow , L.L. , Keeler , C.L. , Tessmer , L.L. , Czymmek , K. and Dohms , J.E. 1998 . Characterization of MGC2, a Mycoplasma gallisepticum cytadhesin with homology to the Mycoplasma pneumoniae 30-kilodalton protein P30 and Mycoplasma genitalium P32 . Infection and Immunity , 66 : 3436 – 3442 .
- Jarosinski , K.W. , Yunis , R. , O'Connell , P.H. , Markowski-Grimsrud , C.J. and Schat , K.A. 2002 . Influence of genetic resistance of the chicken and virulence of Marek's disease virus (MDV) on nitric oxide responses after MDV infection . Avian Diseases , 46 : 636 – 649 .
- Keeler , C.L. Jr , Hnatow , L.L. , Whetzel , P.L. and Dohms , J.E. 1996 . Cloning and characterization of a putative cytadhesin gene (mgc1) from Mycoplasma gallisepticum . Infection and Immunity , 64 : 1541 – 1547 .
- Kleven , S.H. 1998 . In D.E. Swayne , J.R. Glisson , M.W. Jackwood , J.E. Pearson & W.M. Reed Mycoplasmosis. A Laboratory Manual for the Isolation and Identification of Avian Pathogens (pp. 74 – 80 ). Kennett Square , PA : American Association of Avian Pathologists .
- Kollias , G.V. , Sydenstricker , K.V. , Kollias , H.W. , Ley , D.H. , Hosseini , P.R. , Connolly , V. and Dhondt , A.A. 2004 . Experimental infection of house finches with Mycoplasma gallisepticum . Journal of Wildlife Diseases , 40 : 79 – 86 .
- Ley , D.H. , Berkhoff , E. and McLaren , J.M. 1996 . Mycoplasma gallisepticum isolated from house finches (Carpodacus mexicanus) with conjunctivitis . Avian Diseases , 40 : 480 – 483 .
- Ley , D.H. , Sheaffer , D.S. and Dhondt , A.A. 2006 . Further western spread of Mycoplasma gallisepticum infection of house finches . Journal of Wildlife Diseases , 42 : 429 – 431 .
- Li , Y.P. , Bang , D.D. , Handberg , K.J. , Jorgensen , P.H. and Zhang , M.F. 2005 . Evaluation of the suitability of six host genes as internal control in real-time RT-PCR assays in chicken embryo cell cultures infected with infectious bursal disease virus . Veterinary Microbiology , 110 : 155 – 165 .
- Long , E.O. and Dawid , I.B. 1980 . Repeated genes in eukaryotes . Annual Review of Biochemistry , 49 : 727 – 764 .
- Lopez-Andreo , M. , Lugo , L. , Garrido-Pertierra , A. , Prieto , M.I. and Puyet , A. 2005 . Identification and quantification of species in complex DNA mixtures by real-time polymerase chain reaction . Analytical Biochemistry , 339 : 73 – 82 .
- Mekkes , D.R. and Feberwee , A. 2005 . Real-time polymerase chain reaction for the qualitative and quantitative detection of Mycoplasma gallisepticum . Avian Pathology , 34 : 348 – 354 .
- Papazisi , L. , Frasca , S. Jr , Gladd , M. , Liao , X. , Yogev , D. and Geary , S.J. 2002 . GapA and CrmA coexpression is essential for Mycoplasma gallisepticum cytadherence and virulence . Infection and Immunity , 70 : 6839 – 6845 .
- Sydenstricker , K.V. , Dhondt , A.A. , Hawley , D.M. , Jennelle , C.S. , Kollias , H.W. and Kollias , G.V. 2006 . Characterization of experimental Mycoplasma gallisepticum infection in captive house finch flocks . Avian Diseases , 50 : 39 – 44 .
- Vogl , G. , Plaickner , A. , Szathmary , S. , Stipkovits , L. , Rosengarten , R. and Szostak , M.P. 2008 . Mycoplasma gallisepticum invades chicken erythrocytes during infection . Infection and Immunity , 76 : 71 – 77 .
- Weinstock , D. and Schat , K.A. 1987 . “ Virus specific syngeneic killing of reticuloendotheliosis virus transformed cell line target cells by spleen cells ” . In Avian Immunology , Edited by: Weber , W.T. and Ewert , D.L. 253 – 263 . New York : Alan R. Liss .
- Winner , F. , Rosengarten , R. and Citti , C. 2000 . In vitro cell invasion of Mycoplasma gallisepticum . Infection and Immunity , 68 : 4238 – 4244 .