Abstract
Infections with pigeon circovirus (PiCV) (also termed columbid circovirus) occur in meat and racing pigeons (Columba livia) of all ages and have been reported worldwide. A PiCV infection is associated with immunosuppression and the development of young pigeon disease syndrome. An indirect enzyme-linked immunosorbent assay (ELISA) for the detection of virus-specific serum antibody was developed for research purposes. In the absence of a method to propagate PiCV in cell culture, the assay was based on a recombinant truncated capsid protein (rCapPiCV) produced by overexpression in Escherichia coli. A 6×His-Tag was fused to the N-terminus of the protein to facilitate purification by metal affinity chromatography and detection by anti-His antibody. PiCV-negative and PiCV-positive control sera were generated by inoculation of pigeons with tissue homogenate containing PiCV, followed by five weekly blood sample collections. Western blotting of the immune serum revealed a specific protein band of approximately 32 kDa, which was absent in the pre-immune sera. Using rCapPiCV as antigen in an indirect ELISA, PiCV-specific antibody was detected in sera of the experimentally PiCV-infected pigeons collected at 1 to 5 weeks post infection. By testing 118 field sera collected in the years 1989, 1991, 1994 and 2008 in the rCapPiCV ELISA, virus-specific antibody was detected in 89 (75%) of the sera. The results obtained demonstrate that the rCapPiCV-based indirect ELISA is able to detect PiCV-specific antibodies in pigeon sera and may be a useful tool for PiCV serodiagnosis.
Introduction
The first cases of circovirus infection in pigeons (Columba livia) were reported in the US (Shivaprasad et al., Citation1993; Woods et al., Citation1993). Ever since, the agent has been identified in diseased pigeons from South Africa (Gerdes, Citation1993), Australia, Canada (Woods et al., Citation1994), the UK (Smyth & Caroll, Citation1995), Germany (Soike, Citation1997), Italy (Coletti et al., Citation2000), France (Abadie et al., Citation2001) and the Czech Republic (Taras et al., Citation2003). Circoviruses are the smallest animal viruses known. They are characterized by a single-stranded circular DNA genome of approximately 2000 nucleotides. The genus Circovirus includes the members porcine circovirus type 1 (PCV1) and type 2 (PCV2) (Tischer et al., Citation1982; Mankertz et al., Citation1997), psittacine beak and feather disease virus (BFDV) (Bassami et al., Citation1998), pigeon circovirus (PiCV) or columbid circovirus (Mankertz et al., Citation2000), goose circovirus (Soike et al., Citation1999), canary circovirus (Phenix et al., Citation2001) and the tentative members duck circovirus (Hattermann et al., Citation2003), finch circovirus (Shivaprasad et al., Citation2004) and gull circovirus (Twentyman et al., Citation1999; Todd et al., Citation2005). Circoviruses have also been detected in the following avian species, but have not yet been classified as members of the genus Circovirus: ostriches (Struthio camelus) (Eisenberg et al., Citation2003), Australien ravens (Corvus coronoides) (raven circovirus) (Stewart et al., Citation2006) starlings (Sturnus vulgaris and Sturnus unicolor) (starling circovirus) (Johne et al., Citation2006) and mute swans (Cygnus olor) (swan circovirus) (Halami et al., Citation2008). Phylogenetic analysis of avian and mammalian circoviruses and their hosts’ cytochrome b indicate a host-specific evolution of circoviruses without virus transmission from distantly related hosts (Johne et al., Citation2006).
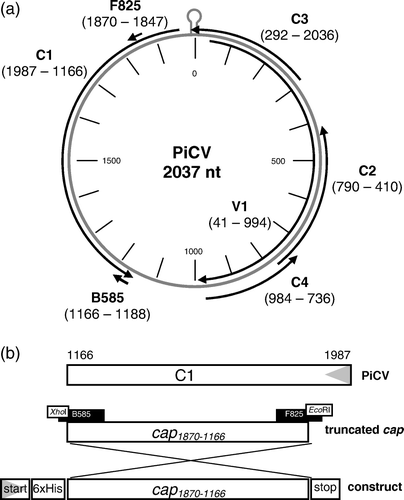
PiCV is characterized by a circular, single-stranded ambisense-organized DNA genome of 2037 nucleotides with five open reading frames (ORFs) (a). The putative capsid protein (Cap) is encoded by ORF C1 on the minus strand of PiCV and has a size of 274 amino acids or 31.9 kDa (positions 1987 to 1166) (Mankertz et al., Citation2000). Avian circoviruses show a distinct tropism to lymphoreticular tissue (Johne et al., Citation2006), the main signs of infection being immunosuppression and feather disorders (reviewed by Todd, Citation2004). In pigeons, PiCV-specific pathological changes are essentially restricted to primary and secondary lymphatic tissues (Soike et al., Citation2001), ranging from mild lymphocytic depletion to cystic bursal atrophy with severe lymphoid depletion (Woods et al., Citation1994; Paré et al., Citation1999; Coletti et al., Citation2000; Soike et al., Citation2001; Raue et al., Citation2005; Scullion & Scullion, Citation2007; Schmidt et al., Citation2008). Therefore it was suggested that PiCV induces a severe immunosuppression in young pigeons, predisposing infected animals to opportunistic secondary infections that would normally be controlled by humoral or cell-mediated immunity (Woods & Shivaprasad, Citation1997). Subsequently PiCV infection was considered a crucial factor for the development of young pigeon disease syndrome (YPDS), a multifactorial disease characterized by a broad range of unspecific clinical signs including lethargy, anorexia, poor racing performance, weight loss, respiratory distress and diarrhoea (Raue et al., Citation2005). However, the role of PiCV for the pathogenesis of YPDS remains uncertain as experimental PiCV infection and simultaneous paramyxovirus serotype 1 (APMV1) vaccination of young pigeons has failed to reproduce YPDS or induce suppression of APMV1 antibody response (Schmidt et al., Citation2008).
Figure 1. 1a: Genome organization of PiCV. Black arrows, transcription of potential ORFs of PiCV, labelled according to their localization on the virus (V) or the complementary (C) strand. Positions of start and stop codons are given by numbers. The stem-loop element represents the putative origin of replication (Mankertz et al., Citation2000). Small arrows, position of the primers used to amplify cap1870–1166. 1b: Schematic presentation of the construction of pTriEx-6HN-cap1870–1166. Grey triangles, direction of transcription. Nucleotide positions are given by numbers. Black boxes, primers with primer encoded adjacent restriction enzymes, used to amplify cap1870–1166.
Although PiCV is described to be the most frequently detected infection in young diseased pigeons (Soike et al., Citation2001), more recent studies demonstrate PiCV infection in apparently healthy adult birds, thus suggesting that many PiCV infections may be subclinical (Paré et al., Citation1999; Todd et al., Citation2002; Franciosini et al., Citation2005; Raue et al., Citation2005; Duchatel et al., Citation2006) and persisting, thereby creating a source of infection for younger birds. Still, little is known about the prevalence of infection, especially in adult and newly hatched pigeons (Duchatel et al., Citation2006). To date, diagnosis of PiCV infection relies on detection of viral DNA by polymerase chain reaction (PCR) (Soike et al., Citation2001; Todd et al., Citation2002, Citation2008; Roy et al., Citation2003; Franciosini et al., Citation2005; Freick et al., Citation2008), dot blot analysis and in situ hybridization (Smyth et al., Citation2001; Todd et al., Citation2002) to confirm clinical and histological findings. However, intra vitam diagnosis of PiCV is difficult, as testing of blood, tracheal, pharyngeal and cloacal swabs failed to detect all birds found to be infected at necropsy (Franciosini et al., Citation2005; Duchatel et al., Citation2006; Todd et al., Citation2006). Therefore, antibody detection is an invaluable tool to evaluate the seroprevalence of PiCV in live wild and captive pigeons, but studies on seroprevalence and antibody response have been hampered by the absence of a culture-based virus propagation system. Expression of recombinant capsid protein represents a suitable alternative of antigen production for detection of PiCV-specific serum antibody.
In this study, recombinant expression of a truncated and His-tagged capsid protein of PiCV (rCapPiCV) in Escherichia coli (E.coli) and its applicability to serological tests is investigated.
Materials and Methods
Construction of plasmid
All plasmids were constructed by cloning of PCR fragments. For the generation of pTriEx-6HN, oligonucleotides F524 (5′-TCATCATCATCACCATCACAGCAGCGGCGAGCT-3′) and B525 (5′-CGCCGCTGCTGTGATGGTGATGATGATGA-3′) were hybridized and cloned into the EcoRV-restricted and SacI-restricted vector pTriEx1.1 (Novagen, Darmstadt, Germany), enabling the expression of N-terminally 6×His-tagged proteins.
For the generation of pTriEx-6HN-cap 1870–1166, PCR fragments were amplified with high-fidelity polymerase (Roche Diagnostics, Mannheim, Germany) from a PiCV-isolate (Ger 1; GenBank accession number AF252610), using primers F825 (5′-CCGAATTCTTACTTCCGCCTACGTCGCAAGGAC-3′) and B585 (5′-GTGCTCGAGTTATTCAGAATCCACAGCTGAGT-3′). These primers contained engineered EcoRI and XhoI sites (underlined) to enable insertion of the amplified PCR product into the vector (b). PiCV-specific primers were selected on the basis of a published PiCV genome (GenBank accession number AF252610; Mankertz et al., Citation2000) to amplify ORF C1 without the 117 N-terminal nucleotides (positions 1870 to 1166), due to their high share of arginine codons (Mankertz et al., Citation2000). The PCR product cap 1870–1166 was cloned into an EcoRI-restricted and XhoI-restricted vector pTriEx-6HN. The resulting vector was sequenced to exclude PCR-acquired misincorporations and designated pTriEx-6HN-cap 1870–1166.
Transformation of E. coli and plasmid preparation
For multiplication, pTriEx-6HN-cap 1870–1166 was transformed into One Shot Top10 Chemically Competent E. coli (Invitrogen, Karlsruhe, Germany) in accordance with the manufacturer's protocol. Three single colonies from agar plates were each inoculated in 4 ml medium. All bacteria cultures were incubated at 37°C, liquid cultures on a shaking platform overnight. Luria–Bertani (LB) medium and agar plates (AppliChem, Darmstadt, Germany) supplemented with ampicillin (50 µg/ml) were used. Plasmids were harvested using the NucleoSpin Plasmid kit (Macherey-Nagel, Düren, Germany) according to the manufacturer's user manual. Plasmid concentration was measured photometrically (Eppendorf, Biophotometer, Cologne, Germany).
Expression of rCapPiCV in E. coli
Then 20 µl competent E. coli (RosettaBlue(DE3)LysS) (Novagen) were transformed with 10 ng purified pTriEx-6HN-cap 1870–1166 according to the manufacturer's protocol. Transformed cells (2.5 µl) were diluted in 100 ml LB-Amp-Cam-Glc medium containing 1% glucose, 50 µg/ml ampicillin and 34 µg/ml chloramphenicol. The overnight culture was diluted 1:20 in 2 l LB-Amp-Cam-Glc and grown to an optical density at a wavelength of 600 nm (OD 600) of 0.6. Protein expression was induced by 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) (Novagen). Bacteria cultures were incubated at 37°C on a shaking platform in LB medium (AppliChem). For analysis of protein expression, 500 µl aliquots before and every 60 min after IPTG induction were centrifuged (16,100×g, 1 min, 4°C) and the cell pellets stored at −20°C. After 5 h, cells were harvested by centrifugation (3000×g, 10 min, 4°C). Additionally, 300 ml transformed RosettaBlue(DE3)LysS were grown to OD 600 of 0.6 without induction by IPTG, lysed by sonication on ice and stored at −20°C.
Purification of rCapPiCV
All purification steps were performed at room temperature. The cells were lysed by incubation in 5 ml buffer B+ (100 mM Na2PO4, 10 mM Tris-Cl, 8 M urea, 15 mM imidazole, pH 8.0) per gram of wet cell pellet for 60 min on a rotary shaker. The lysate was cleared by centrifugation at 1000×g for 30 min, and the supernatant used for purification of His-tagged proteins by metal affinity chromatography.
Ni-nitrilotriacetic acid agarose beads (Ni-NTA-Agarose) (Qiagen, Hilden, Germany) and the cleared lysate (v/v 5:1) were incubated on an rotary shaker for 60 min. The lysate-resin mixture was loaded to a 20 ml column and washed with 1 ml buffer C (100 mM Na2PO4, 10 mM Tris-Cl, 8 M urea, pH 6.3) per millilitre of lysate. Proteins were eluted in two pH steps, using 1 ml/2 ml lysate of buffers D and E (100 mM Na2PO4, 10 mM Tris-Cl, 8 M urea, pH 5.9 and pH 4.5, respectively; buffer E containing 250 mM imidazole). Thirty-microlitre aliquots of lysis and purification steps were stored at −20°C. Based on sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE) analysis, fractions of high-protein concentration were pooled (EL) and the total protein amount was quantified by 2-D Quant Kit protein assay (Amersham Pharmacia, Freiburg, Germany).
SDS-PAGE and western blot
Proteins were separated by 12% SDS-PAGE under denaturating conditions, following standard procedures (Laemmli, Citation1970). As protein standards, the molecular weight marker Page Ruler (Fermentas, St Leon-Rot, Germany) for SDS-PAGE and the prestained markers Prestained Protein-Marker (Peqlab GmbH, Erlangen, Germany) and Prestained Page Ruler (Fermentas) for immunoblotting were used. Proteins were stained with Coomassie Brilliant Blue. For western blot analysis, proteins were transferred to nitrocellulose membranes (Amersham Biosciences, Freiburg, Germany) by semi-dry blotting. The membranes were blocked with blocking buffer (2% w/v skim milk powder in phosphate-buffered saline (pH 7.4) supplemented with 0.1 Tween 20 (PBS-T)) for 1 h, followed by three washings with PBS-T. For analysis of individual sera, the membrane was cut into strips. All incubations were performed on a rocking platform and followed by three washings with PBS-T. The His-tag was detected using a monoclonal anti-His mouse antibody (Penta-His Antibody; Qiagen) diluted 1:2000 in blocking buffer incubated at 4°C overnight, followed by a horseradish peroxidase (HRP)-conjugated goat anti-mouse immunoglobulin (Ig)-specific polyclonal antibody (goat anti-mouse-IgG-HRP; Sigma, Munich, Germany) diluted 1:1000 in blocking buffer incubated for 30 min at room temperature. Pigeon sera were diluted 1:250 in blocking buffer and incubated at 4°C followed by a rabbit-derived pigeon IgG-specific antiserum (rabbit anti-pigeon-IgG) (Grund et al., Citation2001) diluted 1:1000 in blocking buffer and a HRP-conjugated goat anti-rabbit IgG specific antibody (anti-rabbit-IgG-HRP) (Sigma) diluted 1:10,000 in blocking buffer. Antigen conjugated to HRP-labelled antibodies was visualized by autoradiography on medical X-ray film (Agfa Curix HT 1.000 G+) using a HRP-substrate (ECL Plus; Amersham Biosciences) according to the manufacturer's protocol. Randomly selected immune sera were incubated with the lysed uninduced host cell culture for 60 min at room temperature and then processed as described above. The reactivity of those sera was compared with and without pre-incubation with lysed host cells.
Enzyme-linked immunosorbent assay
To determine the optimal antigen concentration and serum dilution, different rCapPiCV concentrations (5, 10, 20 and 40 µg/ml) were tested against two-fold serial dilutions of pigeon serum that tested positive for PiCV antibodies in western blot. Subsequently, the optimal antigen concentration and sera dilution (10 µg/ml and 1:50, respectively) were used.
The enzyme-linked immunosorbent assay (ELISA) was performed in Polysorb Immuno Plates (Nunc, Wiesbaden, Germany). To permit binding of the antigen, plates were incubated overnight at room temperature with 50 µl/well purified rCapPiCV diluted in bicarbonate buffer (0.05 M bicarbonate in ddH2O, pH 9.6). Plates were washed three times in washing buffer (Phosphate-buffered saline plus 0.025% Tween 20) and blocked for 60 min with 100 µl/well blocking buffer (2% w/v skim milk powder in washing buffer) followed by three washing steps. Sera were diluted in blocking buffer and 50 µl/well was transferred in duplicate to the antigen-coated wells. After a 30-min incubation and three subsequent washing steps, 50 µl rabbit-derived pigeon IgG-specific antiserum (rabbit anti-pigeon-IgG) (Grund et al., Citation2001) diluted 1:50 in blocking buffer was added to each well and incubated for 30 min. Plates were then washed three times and incubated for 30 min with a HRP-labelled goat derived anti-rabbit IgG antibody (anti-rabbit-IgG-HRP) (Sigma) diluted 1:500 in blocking buffer. Colour development was accomplished by adding 100 µl/well o-phenylenediamine hydrochloride (1% w/v) dissolved in citrate buffer plus hydrogen peroxide (0.05 M sodium citrate in ddH2O, pH 4+0.15% v/v H2O2). After 10-min incubation in darkness, the reaction was stopped by adding 50 µl/well 2 M sulphuric acid in ddH2O. Optical densities were measured with an automated plate spectrophotometer (MR 5000; Dynatech Laboratories) using a test filter with an absorbtion wavelength of 490 nm with a reference wavelength of 650 nm. Positive reactions were defined as reactions resulting in a mean OD higher than the mean extinction plus the three-fold standard deviation of all pre-immune sera. For titration the sera were diluted in blocking buffer starting at a dilution of 1:50 followed by two-fold serial dilution steps. To allow for background noise, two wells per plate were not incubated with sera, but only blocking buffer, and then processed as described above.
To express reactivity of field sera, OD values of the test sera were normalized by subtracting the OD value of the negative control serum and subsequently the ratio of reactivity of individual serum compared with the positive control serum was calculated (S/P ratio):
Polymerase chain reaction
DNA was prepared using a DNAeasy tissue kit (Qiagen) in accordance with the manufacturer's instructions. Isolated DNA was tested for the presence of PiCV DNA by amplifying a 336-nucleotide product within ORF C1 by nested PCR, applying the protocol described by Soike et al. (Citation2001). Amplificates were separated by gel electrophoresis (2% agarose) and stained with ethidium bromide.
Experimental infection of pigeons with PiCV
Experiments were done according to the German animal welfare regulations and authorized by the Regierung von Oberbayern (AZ.: 209.1/211-2531-63/04). Lymphoid tissue (bursa of Fabricius, spleen, bone marrow) of two pigeons tested positive for PiCV by PCR was homogenized and diluted 1:10 in minimum essential medium (Eagle) (MEM) (Biochrom, Berlin, Germany). For further analysis, 100 µl was removed and the remainder was sterile filtrated (0.45 µm) and stored at −80°C. Fifty microlitres of each was cultivated on chicken embryo fibroblasts and chicken embryo liver cells. After three subsequent weekly passages, the homogenate was determined to be free of chicken embryo fibroblast-culturable and chicken embryo liver-culturable pathogens. The source of the birds used for this trial is a pigeon herpesvirus-1-free flock of pigeons housed under specific pathogen free conditions at the Klinik für Vögel (Messana et al., Citation1997) (LMU, Munich, Germany), which was regularly tested for PiCV, pigeon herpesvirus 1, APMV1, Salmonella spp. and Chlamydophila spp. The flock had never tested positive for any of those pathogens. The pigeons were housed under positive pressure and fed sterilized feed. Personnel leave and enter the facility through an air lock and changeroom. Of that flock, 30 pigeons between 4 and 8 weeks of age were selected, randomly divided into two groups and housed separately in a quarantine ward. The quarantine ward included negative pressure, air lock and individual changerooms. After 1 week of acclimatization, one group was injected with 0.5 ml PiCV-containing MEM intramuscularly (Musculus pectoralis profundus) and the control group was mock-infected with MEM. Serum samples were collected (Vena ulnaris) prior to inoculation followed by weekly sample collection. Blood samples were centrifuged and the sera stored at −20°C. In the infected group, five, two and eight pigeons were submitted for necropsy at 3, 4 and 5 weeks, respectively. All control birds were postmortem examined after 5 weeks. Neither infected nor control birds showed any signs of disease. Lymphatic tissues were collected aseptically and immediately stored at −80°C. PiCV-PCR detected PiCV DNA in all infected birds but in none of the control birds.
Field sera
One hundred and eighteen pigeon serum samples were tested for PiCV-specific antibodies by ELISA. Pre-immune and immune sera of the experimentally PiCV-infected pigeons served as negative and positive controls. Eighty-one sera were diagnostic samples of former patients of the Klinik für Vögel (LMU). Of those, 23 sera were collected in 1989, 38 sera in 1991 and 20 sera in 1994. The samples from 1989 and 1991 include sera from wild, racing and fancy pigeons; the samples from 1994 originate from wild pigeons only. The samples were stored at −20°C. Additionally, serum samples of a breeding flock, whose offspring was tested PiCV-positive by PCR (Todd et al., Citation2006) 6 months before, were collected and tested for PiCV-specific serum antibody by ELISA. The flock consists of 37 meat pigeons housed in two stables with 17 and 20 pigeons each. No pigeons had been added to the flock for several years. The offspring had been removed after the PiCV infection was diagnosed. Neither adult pigeons nor offspring had shown signs of disease.
Results
Cloning and bacterial expression of a truncated PiCV capsid protein
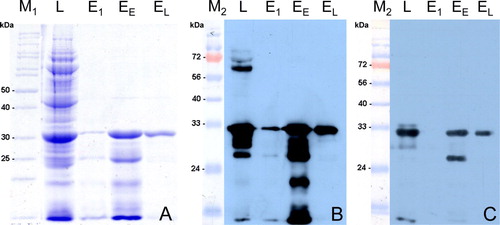
The correct construction of the plasmid was confirmed by sequencing. A plasmid expressing amino acids 40 to 274 of the PiCV Cap protein was generated. In vitro expression and purification of rCapPiCV in E. coli and subsequent SDS-PAGE revealed a highly purified protein with an apparent molecular weight of 32 kDa eluted at pH 4.5 (a). A protein concentration of 0.506 mg/ml was determined for the pooled elution fraction EL.
Figure 2. Protein expression and purification of rCapPiCV. Level of rCapPiCV expression and purification steps were analysed by (2a) Coomassie staining, and western blot analysis using either (2b) an α-His antibody or (2c) sera of experimentally PiCV-infected pigeons. L, cleared lysate; E1, elution at pH 5.9; EE, early elution fractions at pH 4.5; EL, late elution fractions at pH 4.5; M1 and M2, protein markers, respective molecular weights given by numbers.
Antigenicity of rCapPiCV
Since antisera against PiCV are not yet available, an anti-His antibody, and pre-immune and immune sera of experimentally infected pigeons were used to assess antigenicity of rCapPiCV. In western blot analysis a specific protein band of the expected size was detected by the anti-His antibody and the immune serum (b and 2c), but not by the pre-immune serum (data not shown).
Testing of bird sera using purified rCapPiCV by western blotting
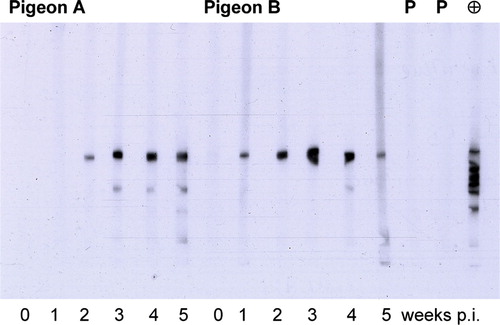
The purified rCapPiCV (EL) was used to detect PiCV-specific serum antibodies by western blot analysis in sera of experimentally PiCV-infected pigeons. In western blot analysis, a specific band of approximately 32 kDa was detected in sera from 12 of the 15 infected pigeons 1 week post infection (p.i.) and by sera from 14 pigeons 2 weeks p.i. By week 3 p.i., all pigeons had detectable amounts of PiCV antibodies. The anti-His antibody detected several additional bands of lower molecular weight. This effect was rarely seen in pigeon sera and only at least 3 weeks p.i. No PiCV-specific antibodies were detected in pre-immune or control group sera. Pre-incubation of immune sera with lysed uninduced host cell culture did not reduce their reactivity, and thereby confirms the absence of interfering E. coli proteins. Cross-reactivity with BFDV antibodies, using sera of two psittacine birds positive for BFDV antibodies by haemagglutination inhibition test, was not observed ().
Figure 3. PiCV-specific reactivity of pigeon sera using purified rCapPiCV (EL) in western blot analysis. Sera of two pigeons at different time points after PiCV infection; in addition, two psittacine birds infected with BFDV were tested (P). As positive control the α-His antibody was included (⊕). Reactivity is shown after an exposure time of 10 sec. p.i., post infection.
Reaction of bird sera in ELISA
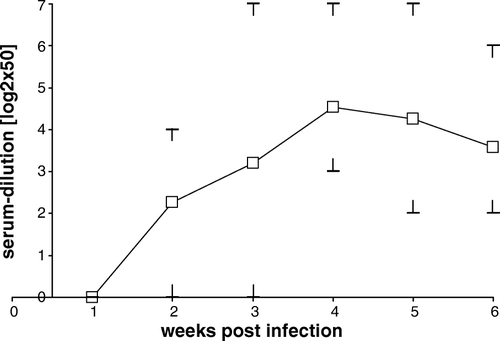
We next analysed the applicability of rCapPiCV in an indirect ELISA system. Pre-immune and immune sera of 15 experimentally infected pigeons were titrated within this test system. The reactivity of the pre-immune sera did not exceed the background noise of rCapPiCV-coated, but of serum-incubated wells. However, 1 week p.i. 12 pigeons had seroconverted and showed titres ranging from 1:50 to 1:400. Analysis of all time points revealed a rise in titres to week 3 p.i. (ranging from 1:200 to 1:3200) and then a slight decline on the fourth and fifth week (titres ranging from 1:100 to 1:3200 and from 1:100 to 1:1600 respectively). Mean, individual minimal and maximal titres at each time point are shown in .
Field sera
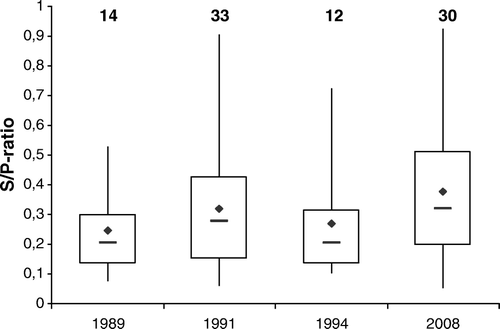
In addition to sera from experimentally infected pigeons, pigeon field sera were tested in the PiCV ELISA. Of 118 sera, 89 tested positive for PiCV antibodies, ranging in reactivity between moderate to distinct (). Of the sera from pigeons collected during the years 1989 and 1991, 14 of 23 sera (61%) and 33 of 38 (87%), respectively, tested positive for PiCV antibodies. Of the sera from feral pigeons collected in 1994, 12 of 20 sera (60%) tested positive. In the serum samples from two breeding flocks, collected in 2008, PiCV antibodies could be detected in 12 of 17 samples (71%) and in 18 of 20 samples (90%). In the serum samples from the breeding flock collected in 2008, PiCV antibodies could be detected in 30 of the 37 samples (81%).
Figure 5. Detection of PiCV-specific antibodies in pigeon field sera by rCapPiCV-ELISA. A total of 118 sera from four different years were tested. The years and the number of positive sera are given by numbers. Reactivity of the sera was standardized as the S/P ratio as described in Materials and Methods, and the box plot is showing the median (bold line), the average (♦), and the lower and upper hinges (25% and 75%, respectively) of the PiCV ELISA-positive sera.
Discussion
PiCV is the most frequently detected infection in diseased young pigeons (Soike et al., Citation2001) and has been identified throughout the world in meat and racing pigeons of all ages (reviewed by Todd, Citation2004). The control of PiCV-induced disease is challenging since (i) pigeons are regularly traded, transported and raced; (ii) wild pigeons host a virus reservoir; and (iii) reliable antemortem diagnosis is not available. Antibody detection is a necessary tool for disease control, especially as subclinically infected birds shed virus (Franciosini et al., Citation2005; Duchatel, et al., Citation2006; Todd et al., Citation2006). Detection of antibodies against PiCV indicates that, even if no viral antigen can be detected in swabs, the bird nonetheless had contact with the infectious agent. Therefore a suitable serological test, such as the rCapPiCV-based indirect ELISA, can help to determine a more accurate PiCV infection rate of pigeon populations compared with use of PCR alone. This offers the possibility of systematic surveillance of flocks and wild pigeons. However, the development of diagnostic methods and vaccines is restrained by PiCV not being cultivable in conventional virus propagation systems. Therefore, a recombinant antigen presents a suitable alternative. As Cap is considered the capsid protein of PiCV, it is likely to induce measurable antibody response in circovirus-infected pigeons, as has been shown for the Cap of BFDV in psittacine birds (Johne et al., Citation2004), goose circovirus in geese (Scott et al., Citation2006) and PCV2 in swine (Nawagitgul et al., Citation2002). As production of recombinant proteins is a fast and efficient method to produce large amounts of antigen that does not require handling of infected tissues, this approach would present the best method to detect PiCV-specific antibodies, assessing the individual PiCV status as well as for epidemiological surveys.
In this study we investigated the potential of a truncated recombinant Cap of PiCV to detect PiCV-specific serum antibodies. Due to the high proportion of the arginine codons AGG and AGA, which are rarely used in E. coli, the first 39 amino acids were not included in the recombinant Cap. But, if the amino-terminus is involved in packaging of viral DNA into the capsid, and thus located within the capsid, as has been proposed for corresponding regions within the capsid proteins of chicken anaemia virus, the porcine circoviruses (Todd et al., Citation2005) and BFDV (Heath et al., Citation2006), the deletion of the region should not reduce the antigenic potential of rCapPiCV. The truncated Cap was expressed in sufficient amounts in E. coli and purified to a high degree using nickel affinity chromatography. A protein band was detected not only by anti-His antiserum but also by sera of experimentally PiCV-infected pigeons. Its apparent size of 32 kDa corresponded to the mass calculated for rCapPiCV. The value of rCapPiCV as antigen for detection of PiCV-specific antibodies was confirmed by ELISA and western blot analysis, demonstrating seroconversion in all 15 experimentally infected pigeons. Additional bands of lower molecular weight were detected by the anti-His antibody, but only by few of the pigeon sera and considered to be fragments of rCapPiCV containing the His-tag and of minor antigenic relevance. Correlation between ELISA and western blot results was 100%. The fact that 12 pigeons reacted with measurable antibody production to PiCV infection as early as 1 week p.i. might have been due to a cross-reactivity of IgY and IgM light chain. Another possible explanation could be that the flock had been exposed to PiCV prior to the animal trial and the experimental PiCV infection acted as a booster to antibody production. However, no antibodies could be detected in the pre-immune sera and no PiCV DNA could be detected in lymphatic tissue of any of the control birds. As the PCR used is based on the ORF C1, where a certain nucleotide diversity between different PiCV isolates exists (Todd et al., Citation2008), it has to be taken into account that more distantly PiCV strains might have escaped detection. Nevertheless, the data from the infection experiment clearly demonstrate, that rCapPiCV is a suitable antigen for detection of PiCV-specific antibodies and represents epitopes of the native Cap of PiCV. Since sequence comparisons of 12 different PiCV isolates revealed amino acid variability within the Cap, antigenic differences may interfere with the overall ability to recognize an infection with a certain PiCV strain (Todd et al., Citation2008). Testing of field sera in the PiCV ELISA indicated a broad diagnostic sensitivity, detecting seropositive pigeons in all examined groups. However, putative antigenic differences of PiCV strains have to be addressed in further studies.
Regarding the serological data from field sera, it was remarkable that PiCV antibodies were present in sera from the years 1989, 1991 and 1994, suggesting that the virus has circulated in German pigeon populations before it was first detected in Germany in 1997 (Soike, Citation1997). In addition the detection of PiCV-specific antibodies in sera from a breeding flock infected with PiCV indicates that PiCV serology could be useful to assess the PiCV status of pigeon flocks. But it has to be taken into account that the ELISA described here has to be validated further. Before it can be applied in routine diagnosis, a larger number of defined sera from different sources have to be tested. Cross-reactivity between anti-BFDV antibody and rCapPiCV was not observed, corresponding with results of Woods et al. (Citation1993). Moreover, the ability of rCapPiCV to induce a protective immune response against PiCV infection in young pigeons awaits further investigation. In swine, a vaccine containing recombinant PCV2-Cap protects against PCV2 challenge, completely inhibiting virus replication (Blanchard et al., Citation2003).
The rCapPiCV presented in this study provides the opportunity to test for PiCV-specific antibody. This tool can be used in further studies to investigate whether there is a correlation between antibody titre, virus shedding and manifestation of disease. These data are necessary for a correct interpretation of serological results and the development of diagnostic strategies. For example, in BFDV infection it is proposed that high antibody titres are an indicator for recent infection and no virus shedding, while chronically diseased birds do not have detectable titres but shed virus (Khalesi et al., Citation2005). Subsequently the combination of virus and antibody detection can provide useful information on the PiCV status of individuals and flocks. In addition, studying antibody response in PiCV-infected animals will provide an insight into PiCV epidemiology.
Acknowledgements
The authors thank Beate Köhler (Klinik für Vögel, LMU Munich, Germany) for providing the BFDV haemagglutination inhibition-positive parrot sera.
Additional information
Notes on contributors
Iris Daum†
† Present address: Lohmann Animal Health GmbH & Co. KG, Heinz-Lohmann-Str. 4, D–27472, Cuxhaven, GermanyChristian Grund‡
‡ Present address: Institute of Diagnostic Virology, Friedrich-Loeffler-Institut, Federal Research Institute for Animal Health, Südufer 10, 17493, Greifswald-Insel Riems, GermanyReferences
- Abadie , J. , Nguyen , F. , Groizeleau , C. , Amenna , N. Fernandez , B. 2001 . Pigeon circovirus infection: pathological observations and suggested pathogenesis . Avian Pathology , 30 : 149 – 158 .
- Bassami , M.R. , Berryman , D. , Wilcox , G.E. and Raidal , S.R. 1998 . Psittacine beak and feather disease virus nucleotide sequence analysis and its relationship to porcine circovirus, plant circoviruses, and chicken anaemia virus . Virology , 249 : 453 – 459 .
- Blanchard , P. , Mahé , D. , Cariolet , R. , Keranflec'h , A. , Baudouard , M.A. Cordioli , P. 2003 . Protection of swine against post-weaning multisystemic wasting syndrome (PMWS) by porcine circovirus type 2 (PCV2) proteins . Vaccine , 21 : 4565 – 4575 .
- Coletti , M. , Franciosini , M.P. , Asdrubali , G. and Passamonti , F. 2000 . Atrophy of the primary lymphoid organs of meat pigeons in Italy associated with circoviruslike particles in the bursa of Fabricius . Avian Diseases , 44 : 454 – 459 .
- Duchatel , J.P. , Todd , D. , Smyth , J.A. , Bustin , J.C. and Vindevogel , H. 2006 . Observations on detection, excretion and transmission of pigeon circovirus in adult, young and embryonic pigeons . Avian Pathology , 35 : 30 – 34 .
- Eisenberg , S.W. , van Asten , A.J. , van Ederen , A.M. and Dorrestein , G.M. 2003 . Detection of circovirus with a polymerase chain reaction in the ostrich (Struthio camelus) on a farm in The Netherlands . Veterinary Microbiology , 95 : 27 – 38 .
- Franciosini , M.P. , Fringuelli , E. , Tarhuni , O. , Guelfi , G. , Todd , D. Casagrande , P.P. 2005 . Development of a polymerase chain reaction-based in vivo method in the diagnosis of subclinical pigeon circovirus infection . Avian Diseases , 49 : 340 – 343 .
- Freick , M. , Müller , H. and Raue , R. 2008 . Rapid detection of pigeon herpesvirus, fowl adenovirus and pigeon circovirus in young racing pigeons by multiplex PCR . Journal of Virological Methods , 148 : 226 – 231 .
- Gerdes , G.H. 1993 . Two very small viruses—a presumptive identification . Journal of the South African Veterinary Association , 64 : 2
- Grund , C. , Gerbermann , H. and Grimm , F. 2001 . “ Prevalence of subclinical paramyxovirus infections in parrots in Germany: investigations of droppings and sera ” . In Proceedings of the 6th European Association of Avian Veterinarians Conference , 208 – 211 . Munich, Germany .
- Halami , M.Y. , Nieper , H. , Müller , H. and Johne , R. 2008 . Detection of a novel circovirus in mute swans (Cygnus olor) by using nested broad-spectrum PCR . Virus Research , 132 : 208 – 212 .
- Hattermann , K. , Schmitt , C. , Soike , D. and Mankertz , A. 2003 . Cloning and sequencing of Duck circovirus (DuCV) . Archives of Virology , 148 : 2471 – 2480 .
- Heath , L. , Williamson , A.L. and Rybicki , E.P. 2006 . The capsid protein of beak and feather disease virus binds to the viral DNA and is responsible for transporting the replication-associated protein into the nucleus . Journal of Virology , 80 : 7219 – 7225 .
- Johne , R. , Raue , R. , Grund , C. , Kaleta , E.F. and Müller , H. 2004 . Recombinant expression of a truncated capsid protein of beak and feather disease virus and its application in serological tests . Avian Pathology , 33 : 328 – 336 .
- Johne , R. , Fernandez-de-Luco , D. , Hofle , U. and Müller , H. 2006 . Genome of a novel circovirus of starlings, amplified by multiply primed rolling-circle amplification . Journal of General Virology , 87 : 1189 – 1195 .
- Khalesi , B. , Bonne , N. , Stewart , M. , Sharp , M. and Raidal , S. 2005 . A comparison of haemagglutination, haemagglutination inhibition and PCR for the detection of psittacine beak and feather disease virus infection and a comparison of isolates obtained from loriids . Journal of General Virology , 86 : 3039 – 3046 .
- Laemmli , U.K. 1970 . Cleavage of structural proteins during the assembly of the head of bacteriophage T4 . Nature , 227 : 680 – 685 .
- Mankertz , A. , Persson , F. , Mankertz , J. , Blaess , G. and Buhk , H.J. 1997 . Mapping and characterization of the origin of DNA replication of porcine circovirus . Journal of Virology , 71 : 2562 – 2566 .
- Mankertz , A. , Hattermann , K. , Ehlers , B. and Soike , D. 2000 . Cloning and sequencing of columbid circovirus (CoCV), a new circovirus from pigeons . Archives of Virology , 145 : 2469 – 2479 .
- Messana , M. , Kösters , J. and Grund , C. 1997 . Studies on reactivation and transmission of pigeon herpes virus (PHV) for raising PHV-free pigeons (Columba livia dom.) . Avian Pathology , 26 : 859 – 864 .
- Nawagitgul , P. , Harms , P.A. , Morozov , I. , Thacker , B.J. Sorden , S.D. 2002 . Modified indirect porcine circovirus (PCV) type 2-based and recombinant capsid protein (ORF2)-based enzyme-linked immunosorbent assays for detection of antibodies to PCV . Clinical and Diagnostic Laboratory Immunology , 9 : 33 – 40 .
- Paré , J.A. , Brash , M.L. , Hunter , D.B. and Hampson , R.J. 1999 . Observations on pigeon circovirus infection in Ontario . The Canadian Veterinary Journal , 40 : 659 – 662 .
- Phenix , K.V. , Weston , J.H. , Ypelaar , I. , Lavazza , A. , Smyth , J.A. Todd , D. 2001 . Nucleotide sequence analysis of a novel circovirus of canaries and its relationship to other members of the genus Circovirus of the family Circoviridae . Journal of General Virology , 82 : 2805 – 2809 .
- Raue , R. , Schmidt , V. , Freick , M. , Reinhardt , B. , Johne , R. Kamphausen , L. 2005 . A disease complex associated with pigeon circovirus infection, young pigeon disease syndrome . Avian Pathology , 34 : 418 – 425 .
- Roy , P. , Dhillon , A.S. , Lauerman , L. and Shivaprasad , H.L. 2003 . Detection of pigeon circovirus by polymerase chain reaction . Avian Diseases , 47 : 218 – 222 .
- Schmidt , V. , Schlömer , J. , Lüken , C. , Johne , R. , Biere , B. Müller , H. 2008 . Experimental infection of domestic pigeons with pigeon circovirus . Avian Diseases , 52 : 380 – 386 .
- Scott , A.N.J. , Beckett , A. , Smyth , J.A. , Ball , N.W. , Palya , V. and Todd , D. 2006 . Serological diagnosis of goose circovirus infections . Avian Pathology , 35 : 495 – 499 .
- Scullion , F.T. and Scullion , M.G. 2007 . Pathologic findings in racing pigeons (Columba livia domestica) with ‘young bird sickness’ . Journal of Avian Medicine and Surgery , 21 : 1 – 7 .
- Shivaprasad , H.L. , Chin , R.P. , Jeffrey , J.S. , Nordhausen , R.W. and Latimer , K.S. 1993 . “ A new viral disease of pigeons? Particles resembling circovirus in the bursa of fabricius ” . In Proceedings of the 42nd Western Poultry Disease Conference , 99 – 100 . CA , , USA : Sacramento .
- Shivaprasad , H.L. , Hill , D. , Todd , D. and Smyth , J.A. 2004 . Circovirus infection in a Gouldian finch (Chloebia gouldiae) . Avian Pathology , 33 : 525 – 529 .
- Smyth , J.A. and Carroll , B.P. 1995 . Circovirus infection in European racing pigeons . Veterinary Record , 136 : 173 – 174 .
- Smyth , J.A. , Weston , J. , Moffett , D.A. and Todd , D. 2001 . Detection of circovirus infection in pigeons by in situ hybridization using cloned DNA probes . Journal of Veterinary Diagnostic Investigation , 13 : 475 – 482 .
- Soike , D. 1997 . Circovirus infections in pigeons . Tierärztliche Praxis , 25 : 52 – 54 .
- Soike , D. , Köhler , B. and Albrecht , K. 1999 . A circovirus-like infection in geese related to a runting syndrome . Avian Pathology , 28 : 199 – 202 .
- Soike , D. , Hattermann , K. , Albrecht , K. , Segalés , J. , Domingo , M. Schmitt , C. 2001 . A diagnostic study on columbid circovirus infection . Avian Pathology , 30 : 605 – 611 .
- Stewart , M.E. , Perry , R. and Raidal , S.R. 2006 . Identification of a novel circovirus in Australian ravens (Corvus coronoides) with feather disease . Avian Pathology , 35 : 86 – 92 .
- Taras , L. , Kubicek , O. , Juranova , R. and Jurajda , V. 2003 . The first demonstration of pigeon circovirus infection in the Czech Republic based on histology and nested PCR . Acta Veterinaria Brno , 72 : 577 – 582 .
- Tischer , I. , Gelderblom , H. , Vettermann , W. and Koch , M.A. 1982 . A very small porcine virus with circular single-stranded DNA . Nature , 295 : 64 – 66 .
- Todd , D. 2004 . Avian circovirus diseases: lessons for the study of PMWS . Veterinary Microbiology , 98 : 169 – 174 .
- Todd , D. , Duchatel , J.P. , Weston , J.H. , Ball , N.W. , Borghmans , B.J. Moffett , D.A. 2002 . Evaluation of polymerase chain reaction and dot blot hybridisation tests in the diagnosis of pigeon circovirus infections . Veterinary Microbiology , 89 : 1 – 16 .
- Todd , D. , Bendinelli , M. , Biagini , P. , Hino , S. , Mankertz , A. Mishiro , S. 2005 . “ Family Circoviridae ” . In Virus Taxonomy Classification and Nomenclature of Viruses, Eighth Report of the International Committee on the Taxonomy of Viruses , Edited by: Fauquet , C.M. , Mayo , M.A. , Maniloff , J. , Desselberger , U. and Ball , L.A. 327 – 330 . San Diego , CA and London : Elsevier Academic Press .
- Todd , D. , Duchatel , J.P. , Bustin , J.C. , Scullion , F.T. , Scullion , M.G. Scott , A.N. 2006 . Detection of pigeon circovirus in cloacal swabs: implications for diagnosis, epidemiology and control . Veterinary Record , 159 : 314 – 317 .
- Todd , D. , Fringuelli , E. , Scott , A.N. , Borghmans , B.J. , Duchatel , J.P. Shivaprasad , H.L. 2008 . Sequence comparison of pigeon circoviruses . Research in Veterinary Science , 84 : 311 – 319 .
- Twentyman , C.M. , Alley , M.R. , Meers , J. , Cooke , M.M. and Duignan , P.J. 1999 . Circovirus-like infection in a southern black-backed gull (Larus dominicanus) . Avian Pathology , 28 : 513 – 516 .
- Woods , L.W. , Latimer , K.S. , Barr , B.C. , Niagro , F.D. , Campagnoli , R.P. Nordhausen , R.W. 1993 . Circovirus-like infection in a pigeon . Journal of Veterinary Diagnostic Investigation , 5 : 609 – 612 .
- Woods , L.W. and Shivaprasad , H.L. 1997 . “ Pigeon circovirus infection ” . In Diseases of Poultry , 10th edn , Edited by: Calnek , B.W. , Barnes , H.J. , Beard , C.W. , McDougald , L.G. and Saif , Y.M. 1050 – 1053 . Ames : Iowa State University Press .
- Woods , L.W. , Latimer , K.S. , Niagro , F.D. , Riddell , C. , Crowley , A.M. Anderson , M.L. 1994 . A retrospective study of circovirus infection in pigeons: nine cases (1986–1993) . Journal of Veterinary Diagnostic Investigation , 6 : 156 – 164 .