Abstract
Twenty infectious bronchitis virus isolates were recovered from broilers and layers in different outbreaks amongst commercial poultry flocks in different geographic regions of Argentina from 2001 to 2008. The viruses were isolated from the tracheas, lungs, and caecal tonsils of birds that were showing respiratory signs. Further analysis based on their nucleotide and amino acid sequences in hypervariable region (HVR) 1 and the intervening sequence including HVRs 1 and 2 (HVR1/2) of the S1 gene was done to determine the genetic relationships among them and reference strains. Five isolates were highly related to the Massachusetts or Connecticut serotypes, indicating the probability of the detection and isolation of vaccine strains. The other Argentinean isolates formed three separate clusters (A, B and C), distant from the vaccine serotypes, with no correlation between the generated clusters and a geographic pattern. These observations could explain the failure of the Massachusetts serotype vaccination programmes to control IBV in these flocks. In addition, the utilization of HVR1/2 and HVR1 sequences resulted in trees with similar topology but the phylogenetic relationships using HVR1/2 nucleotide sequences were better supported by higher bootstrap values. Therefore, the sequences of the HVR1/2 region are recommended for phylogenetic studies.
Introduction
Infectious bronchitis (IB) is an acute, highly contagious viral respiratory disease of chickens characterized by tracheal rales, nasal exudate, coughing and sneezing/snicking. In addition, there are some strains that cause nephritis and also affect the reproductive system of laying hens, causing a decline in egg production and quality (Bisgaard, Citation1976; Cavanagh & Naqi, Citation2003).
The disease is caused by infectious bronchitis virus (IBV), a single-stranded, positive-sense RNA virus of the Coronaviridae family (Lai & Cavanagh, Citation1997). The genome encodes four structural proteins: the spike (S), membrane (M), nucleocapsid (N) and envelope (E) proteins. The S glycoprotein is proteolytically processed into two non-covalently bound polypeptides S1 and S2 (Stern & Sefton, Citation1982). The S protein is responsible for the attachment of the virion to the host cells, and the S1 subunit is involved in the induction of neutralizing, serotype-specific, and haemagglutination-inhibiting antibodies (Cavanagh et al., Citation1988; Koch et al., Citation1990). The S1 subunit is the most variable protein including three hypervariable regions (HVRs), located within amino acids 38 to 67, 91o 141, and 274o 387 (Cavanagh et al., Citation1988; Moore et al., Citation1998).
The control of IB is based on vaccination with attenuated vaccines. Vaccination programmes against IB must be based on the identification of the IBV strains causing the disease in the field (Hofstad, Citation1981) because nucleotide differences between isolates in S1 sequences are sufficient to change the serotype, and there is poor cross-protection between them (Cavanagh & Naqi, Citation2003; Cavanagh et al., Citation2005). Therefore, identification is important for the selection of the most appropriate vaccine to control the outbreak (Farsang et al., Citation2002).
In Argentina, IB is endemic and is controlled by the use of vaccination using mainly vaccines of the Massachusetts strain. It has been observed that outbreaks of infectious bronchitis sometimes still occur in vaccinated flocks, indicating that, most probably, the emerging isolates were of a different serotype to the vaccine strain. Consequently they were able to escape the immunity conferred by vaccination. This is probably due to the constant evolution of IBV generated by point mutations, insertions, deletions, or RNA recombination of the S1 genes (Cavanagh et al., Citation1992). Therefore, typing IBV field strains is a useful tool for the implementation of control measures and for the understanding of the epidemiology and evolution of IBV.
The aim of this study was to obtain IBV field isolates from different outbreaks in commercial poultry flocks in different geographic regions of Argentina from 2001 to 2008, and to analyse them phylogenetically to determine the relationships between them and reference strains.
Materials and Methods
Samples and virus isolation
Samples were obtained from different outbreaks in commercial layers, broilers and breeders with respiratory signs (). All samples were homogenized (20% w/v) with phosphate-buffered saline (pH 7.0 to 7.2) with antibiotics (10,000 IU/ml penicillin, 10,000 µg/ml streptomycin) and centrifuged (1500×g for 15 min). The supernatant was passed through a sterile 0.22-µm polyethersulphone syringe filter (Whatman Inc., Clifton, New Jersey, USA). The filtered supernatant was used for RNA isolation. When direct reverse transcriptase (RT)-polymerase chain reaction (PCR) amplification had been impossible, the filtered supernatants were inoculated into 9-day-old to 11-day-old specified pathogen-free chicken embryos via the allantoic sac.
Table 1. Samples used in this study
RNA isolation, RT and PCR
For RNA extraction, all samples were processed with the QIAamp Viral RNA Mini Kit (QIAGEN, Valencia, California, USA), according to the manufacturer's instructions. RNA was eluted using 50 µl elution buffer and stored at –70°C. Two negative extraction controls were processed along with each group of samples subjected to RNA extraction.
Reverse transcription was performed with M-MLV RT (Promega). Briefly, total RNA was mixed in a 25 µl reaction containing 150 µg random primers hexamers, 1×M-MLV RT buffer, 200 U M-MLV reverse transcriptase, 2 mM dNTPS, and 20 U RNasin (Promega). The RT reaction was at 42°C for 1 h.
The presence of IBV in the samples was initially detected by the amplification of a small portion (380 base pairs) of the S1 gene using primers XCE1+ and XCE3−, as described previously (Adzhar et al., Citation1997).
Each sample that had been positive for the presence of IBV by PCR was used to amplify HVRs 1 and 2 (HVR1/2) of the S1 gene to perform phylogenetic studies. For the amplification of the HVR1/2 fragment, the forward primer C2U (5′-TGGTTGGCATTTACAYGG-3′) described by Wang & Tsai (Citation1996) was used, and a reverse primer was designed within a conserved region of S1 (from nucleotides 703 to 722) corresponding to a reported sequence from accession number M21970 (IBVrev 5′-CCATCTGAAAAATTACCAGT-3′) in order to include both HVRs. The PCR reaction consisted of 35 cycles of denaturation at 94°C for 30 sec, annealing at 45°C for 40 sec, and polymerization at 72°C for 1 min, followed by a final elongation step of 10 min at 72°C. The amplified products were visualized as previously described.
Every reaction was checked for the presence of PCR inhibitors using a PCR that amplified 152 base pairs of the avian β-actin gene. The primers used for this reaction were based upon the sequence obtained from GenBank with accession number L08165 (nucleotides 685 to 836). Primers βactinFw (5′-GAGAAATTGTGCGTGACATCA-3′) and βactinRv (5′-CCTGAACCTCTCATTGCCA-3′) were used in a PCR reaction comprising 45 cycles of denaturation at 94°C for 30 sec, annealing at 65°C for 30 sec, and polymerization at 72°C for 30 sec, followed by a final elongation step of 3 min at 72°C. The amplified products were visualized as previously described.
Sequencing of the S1 gene
PCR-amplified fragments containing HVR1/2 were purified with the QIAquick Spin Miniprep kit (Qiagen). The DNA fragment was finally eluted with 50 µl elution buffer. The purified PCR products were directly sequenced, using 30 ng template per reaction, according to the instructions of the BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, Foster City, California, USA) as described by the manufacturer. Sequencing reactions were run in an ABI PRISM 3730 Genetic Analyzer (Applied Biosystems). Sequences were edited, saved and analysed with BioEdit software (Hall, Citation1999).
Sequence analysis
Argentine IBV sequences and other sequences available from GenBank were aligned using ClustalX software (Thompson et al., Citation1994), and phylogenetic analyses were calculated using the PHYLIP software package (Felsenstein, Citation1989). The HVR1/2 fragment includes nucleotide positions 134 to 701 (HVR1 fragment includes positions 134 to 324) of the IBV H120 strain genome sequence (GenBank accession number M21970). The sequences established in this paper were deposited in GenBank (). The distance matrices were analysed by the neighbour-joining method (Kimura two-parameter model for nucleotidic sequences and the category George/Hunt/Barker model for amino acidic sequences, transition–transversion rate: 2.0) with 1000 bootstrap replicates. Reference sequences used in this study were: DE072 (EU359658); CU-T2 (U49858); ARK-99 (L10384); GAV-92 (U16157); B1648 (X87238); Holte (L18988); GRAY (L18989); JMK (L14070); Italy-02 (AJ457137); USP-11 (DQ492312); USP-05 (DQ492308); UK/167/84 (X58065); D207 (M21969); D274 (X15832); 6/82 (X04723); KB8523 (M21515); H120 (M21970); Beaudette (DQ001342); M41 (AY851295); Connecticut (EU283057); 4/91 (AF093794); FR/L-1450T/05 (EF079118); and NL/L-1449T/04 (EF079116).
Nucleotide and amino acid differences were calculated with Mega 4.0.2 (Tamura et al., Citation2007) and the standard errors were calculated with a 500-replicate bootstrap.
Results
Virus screening and isolation
From all the samples obtained from 2001 to 2008, only 20 cases were positive for IBV by RT-PCR using XCE1+ and XCE3− primers (). All samples studied in this work showed no PCR reaction inhibitors since all partial amplifications of the β-actin gene were successful. In some cases the samples came from flocks vaccinated with Massachusetts serotype strains (M41 or H120). When amplification of S1 fragments was difficult, virus isolation was performed in specified pathogen-free chicken embryos. All samples inoculated produced characteristic lesions of IBV (curled, haemorrhagic embryos) on the first or second passage ().
Phylogenetic analysis
Every RT-PCR-positive case was used to amplify an approximately 609 base pair region in the S1 gene using C2U and IBVrev primers. A phylogenetic tree constructed on the basis of HVR1/2 sequences showed a similar topology to the one constructed using only HRV1 nucleotide sequences (data not shown), generating three clusters of Argentinean isolates, A, B and C (). Bootstrap values observed in the HVR1/2 tree showed higher bootstrap values in most of the nodes in comparison with the one constructed with only HVR1 sequences.
Figure 1. Phylogenetic tree based on the neighbour-joining method for all of the nucleotide sequences analysed. Numbers along the branches refer to bootstrap values≥70%.
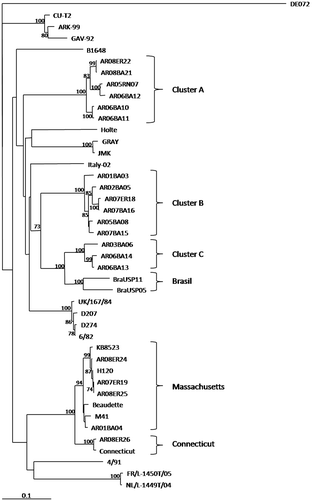
Amino acid and nucleotide differences within groups were equivalent (). In cluster A there are six field isolates clustered separately from South American cluster B and C. In cluster B, there are six field isolates. In cluster C there are three isolates that are closely related to some Brazilian isolates. Although there is an apparent sharing of a common ancestor by groups B and C, partition of them is supported by phylogenetic analysis using both nucleotide and amino acid sequences (data not shown). They can also be clearly distinguished by amino acid differences within the subgroups (6.1±1.1% and 6.3±1.5% of difference for B and C, respectively) and between them (15.5±2.2%).
Table 2. Nucleotide and amino acid similarity (%) within a given groupa
Four isolates grouped with Massachusetts serotype-like isolates. This group has an amino acid difference within their sequences of less than 7% () and of almost 20% with the three Argentinean clusters (). AR08ER26 grouped with another vaccine strain (Connecticut) ().
Table 3. Amino acid relationships between the groupsa
Finally, there was no correlation between the generated groups and any geographic location.
Discussion
In Argentina, commercial flocks are intensely vaccinated against IBV, and these programmes are mostly based on the application of Massachusetts-type vaccines (H120, Ma5, M41, etc.) But, in spite of these intensive vaccination programmes, outbreaks of IB still occur in these commercial flocks. This situation led us to characterize the IBVs circulating in Argentinean flocks, and try to bring more light to the situation in order to help in control this disease.
The finding of four isolates highly related to the Massachusetts serotype indicates the probability of the detection and isolation of vaccine strains. Some of the flocks from where these samples were isolated had been vaccinated with H120 or Ma5 vaccine strains. A similar situation existed with the isolate AR08ER26 but in this case the sequence obtained was phylogenetically grouped with the Connecticut vaccine strain sequence. No information about the vaccination programme of this flock was obtained. It is interesting that these isolates belong to broilers with clinical signs compatible with IB, and in one case (AR/07/ER/19) was co-isolated with an infectious bursal disease virus. Infectious bursal disease virus produces an important immunosuppression, leaving the bird susceptible to co-infections and increasing the clinical signs produced by other agents. Possibly, these immunosuppressed birds developed an enhanced vaccine reaction and showed clinical signs compatible with IB.
The other Argentinean isolates conformed three separate clusters (A, B and C) distinct from the Massachusetts serotype (). Clusters B and C probably belong to a monophyletic group supported by the high bootstrap value observed in the common node, but this observation was not supported when the phylogenetic analysis was made with the amino acidic sequences. This discrepancy opens the possibility of different antigenic and immunogenic behaviour. All of these isolates came from flocks vaccinated with Massachusetts-type vaccines, which produced insufficient protection against these field strains. As noted by Ladman et al. (Citation2006), there is a strongly correlation between the sequence including HVR1/2 of S1 with protective relatedness values, more than antigenic relatedness values based on virus neutralization and haemagglutination inhibition. These observations and the poor relationship between the Argentinean field isolates (from clusters A, B and C) and Massachusetts vaccine strains (average amino acidic identity of 73.6% between clusters A, B and C when compared with the Massachusetts group) could explain the failure of the Massachusetts serotype vaccination programmes to control IBV in these flocks (Gelb et al., Citation2005). Nevertheless, it is necessary to implement a cross-protection analysis between these three clusters and Massachusetts vaccines to establish this failure.
Wang & Tsai (Citation1996) noted that the utilization of the whole S1 nucleotide sequence or the utilization of only HVR1 produced similar relationships, recommending HVR1 for phylogenetic studies. This simplification of the molecular analysis is particularly relevant in some countries that do not have the technology to obtain larger sequences, as, in this case, the whole S1 sequence. In the present work we studied the possibility of expanding this HVR1 region, comparing HVR1 with HVR1/2. The utilization of HVR1/2 and HVR1 resulted in trees with similar topology, but the phylogenetic relationships using HVR1/2 nucleotide sequences brought better support to the majority of the nodes of the tree. All of the cluster nodes were supported by high bootstrap values when HVR1/2 sequences were used () and, in contrast, only some nodes were well supported when only HVR 1 sequences were used (data not shown). The amplification of a PCR product of approximately 600 base pairs is still feasible as a routine PCR and sequencing classification method; therefore, in our opinion, it is recommended to analyse both hypervariable regions instead of only HVR1.
No correlation between the generated groups and a geographic pattern was observed, indicating the high degree of spreading of IBV strains throughout Argentinean commercial flocks. This dispersion is probably due to the constant movement of broilers between different locations; better biosafety conditions must be implemented to control this disease.
This work brings more light on the ecology of IBV in Argentina, but we have to emphasize that a larger sequencing study would be required to reveal the whole spectrum of IBV strains present in Argentinean commercial flocks, and to support the classification of these isolates as a new genotype(s). It is also necessary to study the serological data to complement these findings with the serological relationships between these isolates.
Acknowledgements
This work was supported and financed through Grant projects AESA 3594 (203.940) and AEBIO 2441 (242.410) from the Instituto Nacional de Tecnología Agropecuaria of Argentina. The authors would like to acknowledge the generous support of Andrea Puebla and all of the SIGYSA Team for their technical assistance. They also thank Dr Gustavo Leoni and Dr Mario Plano for providing samples.
References
- Adzhar , A. , Gough , R.E. , Haydon , D. , Shaw , K. , Britton , P. and Cavanagh , D. 1997 . Molecular analysis of the 793/B serotype of infectious bronchitis virus in Great Britain . Avian Pathology , 26 : 625 – 640 .
- Bisgaard , M. 1976 . Infuence of infectious bronchitis virus on egg production, fertility, hatchability and mortality rate in chickens . Nordic Veterinary Medicine , 28 : 368 – 376 .
- Cavanagh , D. and Naqi , S. 2003 . “ Infectious bronchitis ” . In Diseases of Poultry , 11th edn , Edited by: Saif , Y.M. , Barnes , H.J. , Glisson , J.R. , Fadly , A.M. , McDougald , L.R. and Swayne , D.E. 101 – 119 . Ames : Iowa State Press .
- Cavanagh , D. , Davis , P.J. and Cook , J. 1992 . Infectious bronchitis virus: evidence for recombination within the Massachusetts serotype . Avian Pathology , 21 : 401 – 408 .
- Cavanagh , D. , Davis , P.J. and Mockett , A.P. 1988 . Amino acids within hypervariable region 1 of avian coronavirus IBV (Massachusetts serotype) spike glycoprotein are associated with neutralization epitopes . Virus Research , 11 : 141 – 150 .
- Cavanagh , D. , Picault , J.P. , Gough , R. , Hess , M. , Mawditt , K. and Britton , P. 2005 . Variation in the spike protein of the 793/B type of infectious bronchitis virus, in the field and during alternate passage in chickens and embryonated eggs . Avian Pathology , 34 : 20 – 25 .
- Farsang , A. , Ros , C. , Renstrom , L.H. , Baule , C. , Soos , T. and Belak , S. 2002 . Molecular epizootiology of infectious bronchitis virus in Sweden indicating the involvement of a vaccine strain . Avian Pathology , 31 : 229 – 236 .
- Felsenstein , J. 1989 . PHYLIP—Phylogeny Inference Package (Version 3.2) . Cladistics , 5 : 164 – 166 .
- Gelb , J. Jr , Weisman , Y. , Ladman , B.S. and Meir , R. 2005 . S1 gene characteristics and efficacy of vaccination against infectious bronchitis virus field isolates from the United States and Israel (1996 to 2000) . Avian Pathology , 34 : 194 – 203 .
- Hall , T.A. 1999 . BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT . Nucleic Acids Symposium Series , 41 : 95 – 98 .
- Hofstad , M.S. 1981 . Cross-immunity in chickens using seven isolates of avian infectious bronchitis virus . Avian Diseases , 25 : 650 – 654 .
- Koch , G. , Hartog , L. , Kant , A. and van Roozelaar , D.J. 1990 . Antigenic domains on the peplomer protein of avian infectious bronchitis virus: Correlation with biological functions . Journal of General Virology , 71 : 1929 – 1935 .
- Ladman , B.S. , Loupos , A.B. and Gelb , J. Jr . 2006 . Infectious bronchitis virus S1 gene sequence comparison is a better predictor of challenge of immunity in chickens than serotyping by virus neutralization . Avian Pathology , 35 : 127 – 133 .
- Lai , M.M.C. and Cavanagh , D. 1997 . The molecular biology of coronaviruses . Advances in Virus Research , 48 : 1 – 100 .
- Moore , K.M. , Bennett , J.D. , Seal , B.S. and Jackwood , M.W. 1998 . Sequence comparison of avian infectious bronchitis virus S1 glycoproteins of the Florida serotype and five variant isolates from Georgia and California . Virus Genes , 17 : 63 – 83 .
- Stern , D.F. and Sefton , B.M. 1982 . Coronavirus proteins: structure and function of the oligosaccharides of the avian infectious bronchitis virus glycoproteins . Journal of Virology , 44 : 804 – 812 .
- Tamura , K. , Dudley , J. , Nei , M. and Kumar , S. 2007 . MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0 . Molecular Biology and Evolution , 24 : 1596 – 1599 .
- Thompson , J.D. , Higgins , D.G. and Gibson , T.J. 1994 . CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting position – specific gap penalties and weight matrix choice . Nucleic Acid Research , 22 : 4679 – 4680 .
- Wang , C.H. and Tsai , C.T. 1996 . Genetic grouping for the isolates of avian infectious bronchitis virus in Taiwan . Archives of Virology , 141 : 1677 – 1688 .