Abstract
The development of a reverse transcriptase-polymerase chain reaction (RT-PCR) test for detecting chicken astroviruses (CAstV) is described. Primers, which amplified a fragment of 510 base pairs, were located in conserved regions within the ORF 1b (RNA polymerase) gene. The limit of detection of the test was estimated to be approximately 60 viral copies using a 10-fold dilution series of in vitro transcribed RNA. Positive signals were produced with representative CAstV samples, some of which were not detected by a previously described RT-PCR test for detecting CAstV, but other avian astroviruses including avian nephritis virus and duck hepatitis virus types 2 and 3 tested negative. When applied to gut content samples and swabs from UK and German broiler flocks with growth problems, CAstVs were detected by RT-PCR in 50/52 (96%) samples. CAstVs were detected in between 30% and 72.5% pooled gut content samples from longitudinal surveys of four broiler flocks displaying below-average performances. Whereas all day 0 samples were CAstV-negative, high detection rates were observed when the surveyed birds were aged 4, 5 and 7 days. Based on partial ORF 1b sequences, a phylogenetic analysis of 20 CAstVs indicated the existence of two groups. One group comprised four CAstV isolates, including FP3 and 11672, and two field CAstVs, which shared >94% nucleotide identity. The remaining 14 CAstVs, comprising the first characterized CAstV and 612 isolates and 12 field CAstVs, shared 85% to 99% nucleotide identity and displayed 76% to 79% nucleotide identity with the 11672-like and FP3-like CAstVs.
Introduction
Astroviruses are small, spherical, non-enveloped, positive-sense RNA viruses (28 to 30 nm) that cause enteric diseases in mammalian and avian species (Méndez & Arias, Citation2007). Astroviruses of chickens, some of which were originally known as enterovirus-like viruses (McNulty et al., Citation1990; McNeilly et al., Citation1994; Todd et al., Citation2009a), have been implicated in growth depression problems, including runting stunting syndrome (McNulty et al., Citation1984; Spackman et al., Citation1984; Decaesstecker et al., Citation1986; Smyth et al., Citation2007). Runting stunting syndrome is a global problem affecting broiler chicken production, which results in financial losses from increased culling, poor feed conversion and lower uniformity at slaughter with concomitant increased costs from treatment (de Wit, Citation2008).
To date, two different astroviruses have been detected in chickens. Avian nephritis virus (ANV), originally regarded as a picornavirus, was characterized as the first astrovirus of chickens in 2000 (Imada et al., Citation2000). Baxendale & Mebatsion (Citation2004) described the characterization of a novel astrovirus named chicken astrovirus (CAstV) that originated from broiler chicks with runting problems in the Netherlands. CAstVs, which shared high levels of nucleotide sequence identity with the first characterized CAstV (Baxendale & Mebatsion, Citation2004), hereafter known as the reference CAstV, were also detected in US broiler chicken flocks affected by growth problems (Pantin-Jackwood et al., Citation2006). More recently, we have reported the identification of antigenically different CAstV isolates (Todd et al., Citation2009a). Thus, whereas isolate 612, originally described by McNeilly et al. (Citation1994), shared close antigenic and sequence similarities with the reference CAstV, isolates FP3 and 11672 were closely related to one another and more distantly related to the reference CAstV and 612 isolates in terms of antigenicity and partial ORF 1b sequence (Todd et al., Citation2009a).
Establishing the importance of astrovirus infections in broiler growth problems has been difficult due to the absence of convenient diagnostic tests. Electron microscopy is one of the principal means of demonstrating avian astroviruses in diagnostic samples. However, this method relies on observing the star-like morphology (Madeley & Cosgrove, Citation1975), which is not apparent in some types of astrovirus including ANV. In addition, electron microscopy is not suited to high sample throughput and lacks sensitivity (Koci et al., Citation2000; Tang et al., Citation2004; Pantin-Jackwood et al., Citation2007). Isolating astroviruses in cell culture presents difficulties as they grow poorly and are often outgrown by reoviruses and adenoviruses, which also occur commonly in enteric samples. Antigen-detecting diagnostic tests including fluorescent antibody tests performed with cryostat tissue sections or tissue impression smears and antigen capture enzyme-linked immunosorbent assay have not been developed for CAstVs due to the absence of virus-specific antisera.
The detection of CAstVs by reverse transcriptase-polymerase chain reaction (RT-PCR) is a developing field that has been limited both by the low availability of sequence information and the high degree of sequence diversity often displayed by RNA viruses. Degenerate RT-PCR primers, designed by Tang et al. (Citation2004) and based on the avian astrovirus RNA polymerase (pol) gene, were used to amplify ANVs and CAstVs of chicken origin in a US study (Pantin-Jackwood et al., Citation2006). Sequence comparisons of the US CAstV ORF 1b amplicons generated from field samples using these primers were considered when designing the degenerate primers (CAS pol 1F, CAS pol 1R) that were used in the RT-PCR test for detecting CAstV RNA, as described by Day et al. (Citation2007). When used in a multiplex format in which ANV and avian rotaviruses were also detected, this test successfully detected CAstV in 21/34 (62%) surveyed samples from commercial chickens. Recently, we have shown that the ORF 1b sequences of the FP3 and 11672 CAstV isolates differed substantially from those of the USA CAstVs used for designing the CAS pol primer set (Todd et al., Citation2009a). This alerted us to the possibility that the RT-PCR test described by Day et al. (Citation2007) may not be capable of detecting CAstVs with ORF 1b sequences closely resembling the 11672 and FP3 CAstVs.
In this paper we report the development of an RT-PCR test that is capable of detecting both groups of CAstVs. It also describes the application of the test to field samples from broiler flocks with growth problems and from longitudinal surveys of commercial flocks.
Materials and Methods
Viruses and virus growth
The FP3 (Spackman et al., Citation1984) and 612 (McNeilly et al., Citation1994) isolates of CAstV were originally regarded as enterovirus-like viruses. The 11672, 11522 and 1009 CAstV isolates were isolated by yolk-sac inoculation of 6-day-old to 7 day-old chick embryos from weak chicks that were submitted as part of an investigation into hatchability problems (D. Todd, unpublished results). The P22-18.8.00 isolate of CAstV, hereafter named the reference CAstV isolate, was originally isolated by Baxendale & Mebatsion (Citation2004) and was obtained from Dr I. Tarpey, Intervet, Milton Keynes, UK. Information relating to the source, propagation history and production of virus pools of the FP3, 612, 11672, DHV-2 (isolate M52), DHV-3 (isolate X1222A Calnek) and reference (P22 18.8.00) CAstV isolates and the G4260 isolate of avian nephritis virus serotype 1 (ANV-1) used in this investigation was described by Todd et al. (Citation2009a). The 11522 and 1009 CAstV isolates were produced following five and three passages in chick embryos, respectively, using yolk-sac inoculation of homogenates of embryo liver or whole embryo. Subsequently, 10% clarified whole embryo or embryo liver homogenates were used for RNA extraction.
Sample origin and processing
Intestinal contents or cloacal swabs were sampled from chickens collected from problem flocks in the UK and Germany and were stored at −80°C. Samples from the UK were kindly provided by A. Fernadez-Gutierrez (Aviagen Ltd, Midlothian, UK), M. Alcorn (St Davids Poultry Team Ltd, Exmouth, UK), B. H. Thorp (St Davids Poultry Team at the Dick Vet, Easter Bush Veterinary Centre, Midlothian, UK), C. Prins, G. Hayes and A. Walker (Slate Hall Veterinary Practice Ltd, Cambridge, UK), P.W. Cargill (Wyatt Poultry Veterinary Services, Hereford, UK), and D. Pearson (VION UK Veterinary and Food Diagnostic Service, Grampian Country Chickens (Rearing) Ltd, West Lothian, UK). Dr P. Otto (Federal Research Institute for Animal Health, Jena, Germany) provided samples 03V00358, 06V0040 and 06V0062.
Swabs were immersed in 0.5 ml chilled phosphate-buffered saline and vortexed thoroughly. The swab suspensions underwent one freeze/thaw cycle at −80°C before the swab was removed. Intestinal contents were diluted 1:10 in chilled phosphate-buffered saline and then shaken by hand briefly with glass beads to disrupt the solid material. Both suspensions were centrifuged at 3000×g at 4°C for 30 min, and the supernatants were transferred to fresh tubes for storage at −80°C until required. presents the samples investigated.
Table 1. CAstV isolates and field samples examined in the present study
RNA extraction
Viral RNA was extracted from 140 µl each supernatant using the QIAamp Viral RNA Mini Kit (Qiagen, Crawley, UK) according to the manufacturer's instructions. Each RNA was eluted in 30 µl RNase-free water.
RT-PCR assay and direct sequencing
A pair of primers was designed to amplify a 510 base pair (bp) product from CAstVs. The degenerate forward primer, CAstV-for (5′-KCA TGG CTY CAC CGY AAD CA-3′), was located within ORF 1b while the non-degenerate reverse primer, CAstV-rev (5′-CGG TCC ATC CCT CTA CCA GAT TT-3′), was sited in a conserved region located close to the junction of ORF 1b and ORF 2. One-step RT-PCR was performed on the RNAs using the above primers and the SuperScript III One-Step RT-PCR System with the Platinum® Taq DNA Polymerase kit (Invitrogen, Paisley, UK). Each reaction contained 1×reaction buffer, 1 µM each primer, 1 µl enzyme mix, 2.5 µl RNA and diethyl pyrocarbonate-treated H2O to 25 µl. Amplification occurred in a Veriti thermocycler (Applied Biosystems, Warrington, UK), starting with a reverse transcription step of 45°C for 30 min, then an initial denaturing step of 95°C for 2 min, followed by 40 cycles of denaturation at 95°C for 30 sec, annealing at 59°C for 30 sec and extension at 68°C for 30 sec. There was a final extension step at 68°C for 7 min. The CAS pol 1F and CAS pol 1R primers, part of a multiplex RT-PCR assay for detecting CAstV (Day et al., Citation2007), were used in one-step RT-PCR as above with a 55°C annealing temperature and an extension step of 1 min per cycle.
The PCR products were electrophoresed at 125 V for 40 min on a 1% agarose gel in 1×Tris-acetate ethylenediamine tetraacetic acid buffer and were visualized by ethidium bromide staining and ultraviolet transillumination. Amplicons of 510 bp were excised from the gel and purified using the Wizard SV Gel and PCR Clean-Up System (Promega, Southampton, UK). Twenty of the purified PCR products were sequenced in both directions using the same forward and reverse primers used to generate them and the BigDye® Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems). The products were sequenced commercially. Sequencing data were analysed using the Vector NTI suite (Invitrogen). Phylogenetic analysis was conducted on trimmed amplicons of 458 nucleotides using MEGA version 4 (Tamura et al., Citation2007).
Previously described degenerate avian astrovirus primers that amplify a 434 bp fragment from within the pol gene of a wide range of avian astroviruses were used in one-step RT-PCR under previously described conditions to verify the presence of specific astroviruses in ANV, DHV-2 and DHV-3 isolates (Todd et al., Citation2009a). These amplicons were electrophoresed, gel excised, purified and sequenced as described above.
Determination of assay sensitivity
A PCR product of 858 bp, derived from CAstV isolate 11672, was cloned into the pCR®II-TOPO vector (Invitrogen). The PCR product amplified by the forward primer (5′-AGC CTC AAA TAT AGA GCA G-3′) and the reverse primer (5′-GAT CTA GAT GGG GTT TTC TTA G-3′) encompassed 709 bp in ORF 1b and 149 bp in ORF 2. The vector construct was linearized by Xho1 digestion and in vitro transcribed from the SP6 promoter using the MEGAscript® SP6 kit (Ambion, Huntingdon, UK), according to the manufacturer's instructions, but with an overnight incubation period at 37°C. Following transcription, the in vitro transcribed RNA was treated with Turbo DNase (Ambion) and then further purified from template contamination by TRIzol (Invitrogen) extraction. The quality of the in vitro transcribed RNA was assessed by RT-PCR and its purity by PCR using the CAstV-for and CAstV-rev primers and the cycling conditions above (excluding the RT step).
The concentration of the in vitro transcribed RNA was determined using a Genova spectrophotometer (Jenway, Dunmow, UK). The minimum amount of RNA that could be detected by the assay was determined by performing the CAstV RT-PCR on 10-fold serial dilutions of the in vitro transcribed RNA. The products were visualized under ultraviolet transillumination after electrophoresis on a 1% agarose gel stained with ethidium bromide in 1×Tris-acetate ethylenediamine tetraacetic acid buffer. The gene copy limit of detection (LOD) was then calculated for the last dilution visible on the gel.
Longitudinal surveys
Four flocks were screened for the presence of CAstV using RT-PCR based on the CAstV-for and CAstV-rev primers. These flocks were from different sites belonging to the same UK poultry organization and, based on recent performances, were predicted to exhibit below-average performances. Gut contents from ∼12 birds were sampled from each flock at each of 10 timepoints ranging from day 0 to day 42. The 12 samples were grouped into four pools and processed as described above. The performance of each flock was estimated after slaughter by calculating European production efficiency factor (EPEF) values, which represent standard measures of overall flock performance as determined by the equation:
Sequence accession numbers
GenBank accession numbers for the amplicons submitted through this study include: CAstV1009, FJ476291; CAstV11522, FJ476292; CAstVFP3, FJ476293; CAstV612, FJ476294; CAstVP22-08.8.00, FJ476295; CAstVVF04-01/2, FJ476296; CAstVVF04-01/11, FJ476297; CAstVVF05-01/16, FJ476298; CAstV06V0062, FJ476299; CAstVVF06-01/3, FJ476300; CAstVVF06-01/4, FJ476301; CAstVVF06-07/6, FJ476302; CAstVVF06-07/8, FJ476303; CAstVVF07-04/1, FJ476304; CAstVVF07-04/2, FJ476305; CAstVVF07-13/5, FJ476306; CAstVVF08-05/5, FJ476307; CAstVVF08-07/1, FJ476308; and CAstVVF08-05/13, FJ476309.
Results
Location of primers
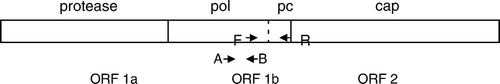
Preliminary analyses identified regions of conserved or semi-conserved sequence suitable for primer design that were shared when 13 representative US CAstV isolates (Pantin-Jackwood et al., Citation2006) and the 11672, FP3 and 612 isolates were compared (unpublished results). The forward primer was sited in the pol gene at a position equivalent to residues 4058 to 4077 in the ANV-1 genome, and contains four degenerate bases. The non-degenerate reverse primer was located 510 bp downstream in a conserved region, that we have termed the precapsid region, located at the 3′ end of the pol gene and including the intergenic region between the pol and cap genes ().
Detection limit of the CAstV RT-PCR test
The LOD of the RT-PCR assay was determined using RNA that had been in vitro transcribed from the recombinant plasmid containing an 858 bp insert generated with the 11672 CAstV isolate. The last 10-fold serial dilution with which a visible RT-PCR product was amplified corresponded to approximately 60 molecules of the viral RNA target (data not shown).
Specificity and performance of CAstV RT-PCR tests
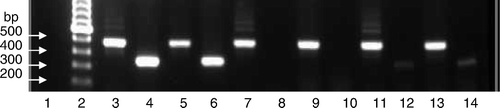
The performance of the RT-PCR tests based on CAstV-for and CAstV-rev primers and the CAS pol 1F and CAS pol 1R primers, used by Day et al. (Citation2007), were compared using representative isolates of CAstV (). Fragments of 510 bp were amplified from the 612, 11672, FP3, 11522 and 1009 isolates and the reference CAstV isolates using the CAstV-for and CAstV-rev primers. However, using the CAS pol 1F and CAS pol 1R primer set, although strongly stained RT-PCR products of 362 bp were generated with the 612 and reference CAstV isolates, very weak signals were obtained using the 11522 and 1009 isolates, and no products were generated from the 11672 and FP3 isolates.
Figure 2. Comparison of CAstV RT-PCR tests performed with different primer sets using cultured CAstV isolates. Upper row, amplicons of 510 bp generated using the CAstV-for and CAstV-rev primers (A). Lower row, amplicons of 362 bp generated using the CAS pol 1F and CAS pol 1R primers (B). Lane 1, negative control; lane 2, molecular weight marker; lane 3, 612 (A); lane 4, 612 (B); lane 5, P22-18.8.00 (A); lane 6, P22-18.8.00 (B); lane 7, 11672 (A); lane 8, 11672 (B); lane 9, FP3 (A); lane 10, FP3 (B); lane 11, 11522 (A); lane 12, 11522 (B); lane 13, 1009 (A); lane 14, 1009 (B).
The specificity of the RT-PCR test based on the CAstV-for and CAstV-rev primers was evaluated using RNAs extracted from ANV, DHV-2 and DHV-3 isolates. Astrovirus-specific fragments of 434 bp were amplified from these RNAs using RT-PCR based on previously described degenerate primers (Todd et al., Citation2009a). However, no amplicons were produced with these RNAs using RT-PCR based on the CAstV-for and CAstV-rev primers.
Detection of CAstVs in field samples
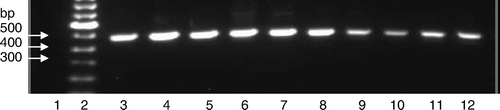
The RT-PCR test based on the CAstV-for and CAstV-rev primers was applied to RNAs that had been extracted from field samples. These were obtained from broiler flocks with enteritis and growth retardation problems, and included 39 gut content and 10 swab samples collected from 10 outbreaks that occurred in five UK poultry organizations from October 2004 to March 2008. Where known, the birds sampled were aged from 4 to 42 days (). Three field samples submitted from problem flocks in Germany from 2004 to 2006 were also investigated. Analysis of the reaction products by agarose gel electrophoresisis followed by ethidium bromide staining and ultraviolet transillumination showed that DNA amplicons of ∼510 bp were clearly visible in 50 of the 52 (96%) field samples tested. In most cases, and with the exception of the primer band, no other DNA bands were visible (). When the RT-PCR test based on the CAS pol 1F and CAS pol 1R primers was applied to the same 52 field samples, 30 (58%) produced amplicons.
Figure 3. Amplification of field sample RNAs by RT-PCR using the CAstV-for and CAstV-rev primers. Lane 1, negative control; lane 2, molecular weight marker; lane 3, VF07-05/3; lane 4, VF07-05/6, lane 5, VF07-14/4; lane 6, VF06-01/3; lane 7, VF06-07/1; lane 8, VF07-13/7, lane 9, VF05-01/3; lane 10, VF05-01/14; lane 11, VF04-01/2 and lane 12, VF04-01/11.
Detection of CAstVs in longitudinal survey samples
CAstVs were detected by RT-PCR using the CAstV-for and CAstV-rev primers in pooled gut content samples from the four broiler flocks () at time points ranging from day 4 to day 42, but all samples were negative for CAstV at day 0. Flocks A and C were CAstV-negative at one-half of the timepoints, with CAstV being detected in 33.3% and 30.0% of the samples in Flocks A and C, respectively. With Flocks B and D, CAstVs were detected at more timepoints (Flock B, eight timepoints; Flock D, nine timepoints) and in higher numbers of pooled samples (Flock B, 72.5%; Flock D, 67.5%). Highest CAstV detection levels across all flocks occurred in samples at days 4 or 5 (14/16, 87.5%), day 7 (12/16, 75%) and day 35 (12/16, 75%). The three male flocks had EPEF flock performance values of 327 (Flock A), 315 (Flock C) and 308 (Flock B), while Flock D, a female flock, had an EPEF value of 238.
Table 2. CAstV detections in longitudinal survey samples from four broiler flocks
Sequence characterization of RT-PCR amplicons
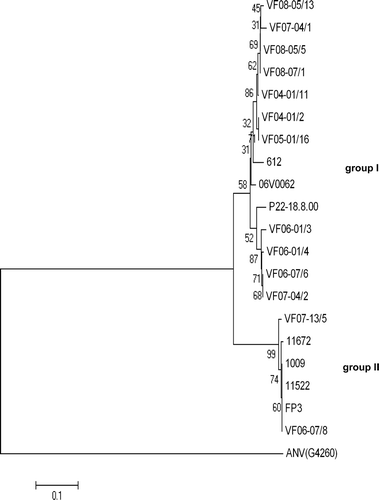
Of the 50 positive samples amplified by the CAstV-for and CAstV-rev primers, 14 field samples were sequenced to confirm CAstV specificity and for characterization purposes. The sequences of the 14 field CAstVs were compared with those from six isolates of CAstV, including 11672, FP3, 1009, 11522, 612 and the reference CAstV isolate, all of which were grown in chick embryos or cell cultures. Phylogenetic analysis showed two major groupings (). One group (Group I), which comprised the 612 and reference CAstV isolates and 12 of the field CAstVs investigated, contained a small number of subgroupings. The second group (Group II) comprised the 11672, FP3, 1009 and 11522 isolates together with the VF06-07/8 and VF07-13/5 field samples. Overall, the 20 CAstVs showed 76% to 99% nucleotide and 81% to 100% amino acid identity over the 458 nucleotide ORF 1b sequences compared. The CAstVs in Group II were closely related, showing 95% to 99% nucleotide and 98% to 100% amino acid identities. In comparison, the 14 CAstVs in Group I were more diverse, showing 85% to 99% nucleotide and 93% to 100% amino acid identity. The group II CAstVs showed 76% to 79% nucleotide and 81% to 85% amino acid identity with the CAstVs comprising Group I.
Figure 4. Phylogenetic tree of CAstV ORF 1b amino acid sequence from selected field samples and isolates. The tree was constructed with Mega 4 (Tamura et al., Citation2007) using neighbour-joining and 1000 bootstrap replicates (bootstrap values are shown on tree). Isolate details are presented in .
Discussion
The present paper describes the development of a sensitive RT-PCR test for detecting CAstV RNA. The high sensitivity of the test, which has an estimated LOD of approximately 60 genome copies, is reflected by the successful detection of CAstV RNA in 50/52 (96%) field samples from broiler flocks with enteritis and growth retardation problems and in 6/6 CAstV isolates that can be propagated in vitro. The test represents an improvement on the test described previously (Day et al., 2007), which, while having a similar estimated LOD, failed to detect CAstVs in 22/52 (42%) field samples and swabs, 50 of which were positive using our newly-described test. Failure to detect CAstV RNA in some of these samples may be due to the inability of the CAS pol primers to amplify ORF 1b sequences that are similar to the FP3 and 11672 isolates, as demonstrated in . The degenerate CAS pol primers were designed following comparison of ∼13 USA CAstV ORF 1b amplicon sequences (Day et al., Citation2007), none of which were 11672-like (Todd et al., Citation2009a). As a consequence, the CAS pol primers were not designed to hybridize to CAstV sequences that resemble those of the FP3 and 11672 isolates. When compared with the sequences of the FP3 and 11672 isolates, analyses showed the presence of several mismatches in both the forward and reverse CAS pol primers, and these are likely to prevent amplification. In contrast, the primers used in the present paper were designed following consideration of both the US CAstV sequences (Pantin-Jackwood et al., Citation2006) and the sequences of 11672, FP3 and 612 CAstV isolates (V. J. Smyth & D. Todd, unpublished results). The forward primer is located within the 434-nucleotide astrovirus ORF 1b amplicon investigated in an earlier study (Todd et al., Citation2009a), while the reverse primer is located in a highly conserved region close to the junction of the ORF 1b and ORF 2. The high degree of sequence conservation displayed by this precapsid region may be due to the role that this part of the genome has as a promoter for the synthesis of the subgenomic RNA that encodes the capsid protein gene.
Based on the partial ORF 1b sequences obtained in this and earlier studies (Pantin-Jackwood et al., Citation2006; Todd et al., Citation2009a), two CAstV groups can be distinguished (). Thus CAstVs with ORF 1b sequences closely similar (>95% nucleotide identity) to the FP3 and 11672 isolates are assigned to Group II, and the CAstVs with ORF 1b sequences that are substantially different (76% to 79% nucleotide identity) are assigned to Group I. However, it would be premature to subdivide CAstVs on the basis of ORF 1b differences alone. Recent work by Strain et al. (Citation2008) involving the analyses of nine complete turkey astrovirus genomes provided evidence for recombination between astrovirus genomes, and supported the view that sequences from different regions of the astrovirus genome should be considered for phylogenetic analyses on which taxonomic subdivisions might be based. With recombination, it is possible that different ORF 1b CAstV sequences could be contiguous with very similar ORF 2 capsid protein sequences, or, conversely, that CAstVs with very similar ORF 1b sequences may have different ORF 2 sequences.
The capsid protein gene is the most diverse astrovirus gene, containing sequence differences that will reflect variation in antigenicity and possibly pathogenicity. Using indirect immunofluorescence, we have recently shown that the FP3 and 11672 isolates of CAstV exhibit low cross-reactivity with the 612 and reference CAstV isolates, suggesting that differences exist in the capsid protein of these isolates. Additional work is needed to determine whether the CAstVs that can be assigned to Group II on the basis of their ORF 1b sequences () also possess distinctive ORF 1a (viral protease) and ORF 2 sequences and distinctive biological properties.
Four of the CAstVs assigned to Group II are isolates that were obtained following inoculation of chick embryos using the yolk-sac route. The FP3 CAstV was originally isolated from dead-in-shell chicks during an investigation of early broiler mortality in the UK (Spackman et al., Citation1984), while isolates 11672, 11522 and 1009 were obtained from weak 1-day-old chicks or late-dead embryos during an investigation of hatchability problems (D. Todd, unpublished results). Together these findings suggest that CAstVs such as the FP3 and 11672 isolates can be vertically transmitted. In comparative pathogenesis experiments involving the inoculation of 1-day-old specific-pathogen-free chicks, the FP3 CAstV isolate was shown to cause greater weight reductions than the 612 CAstV isolate and the G4260 serotype 1 isolate of ANV (Smyth et al., Citation2007). The detection of histological lesions and virus antigen in the kidney and pancreas of FP3-infected chickens indicated that this CAstV isolate can invade beyond the intestine, and this may be a feature of all or some CAstVs. Of the 14 CAstV amplicons generated with field samples that were sequenced in this study, two were closely related to the FP3 and 11672 isolates (Group II; ) and 12 were clustered with the 612 and reference CAstV isolates (Group I; ). With such a small sample size, it would be unwise to draw conclusions about the relative prevalence of the Group I and Group II CAstVs. However, it would appear that more Group I CAstVs were amplified than those belonging to Group II. This may be because the Group I CAstVs are more prevalent, occur in greater amounts when representatives of both groups are present in the same sample, or have RNAs that are more efficiently amplified using the primers.
The availability of an RT-PCR test that can sensitively detect CAstVs provides a useful diagnostic tool with which to begin investigations into CAstV epidemiology and pathogenicity. In the absence of data from healthy flocks, the high detection rate (96%) obtained in this study with samples from growth problem flocks is of uncertain significance. Studies with turkey poults have shown that, in addition to being detected in ∼80% of birds with enteritis and/or growth problems, turkey astroviruses were also detected in ∼30% of birds from “normal” flocks of similar age (Reynolds et al., Citation1987). Therefore, more work is needed to determine the prevalence of CAstV infections in flocks that are unaffected by severe growth retardation problems. In our study, we have tested pooled gut content samples from longitudinal surveys of four broiler flocks, which, in terms of their EPEF performance values, generally displayed below-average performances. The overall CAstV detection levels (30.0% to 72.5%) obtained for the four flocks using pooled samples were lower than that (96%) detected with samples from growth problem flocks (). It was noteworthy that the flocks with the higher EPEF values (Flocks A and C) had lower CAstV detection rates (33.3% and 30.0%) than those of Flock B (72.5%) and Flock D (67.5%), which had lower EPEF values (). However, substantially more flocks, including top-performing flocks (EPEF values >350 for male flocks), would need to be investigated to determine whether CAstV detections are related to broiler performance.
Despite the small number of flocks surveyed, a number of findings were worthy of comment. The lack of detectable CAstV in the day 0 samples and the high detection rates at day 4 or 5 (14/16, 87.5%) and day 7 (12/16, 75.0%) suggested that most birds were infected at very early ages, before day 4. It seems likely that these chicks were infected with horizontally-acquired virus that was either carried over within the house from earlier broiler flocks or excreted by small numbers of vertically infected chicks. We have recently provided serological evidence that CAstV infections are highly prevalent in broiler flocks (Todd et al., Citation2009b). As such, the age at which the majority of chicks are exposed to a substantial CAstV challenge may have an important influence on any adverse effect this astrovirus may have on broiler performance. It remains to be determined whether the CAstVs, which were detected in smaller numbers of samples at later timepoints (for example days 11 to 28), are the same as those that infected the very young chicks and that have continued to be excreted, or whether some birds have become infected with an antigenically different CAstV strain. Infections at later time points might be expected to have less deleterious effects on chicken growth than those at early timepoints.
The severity of the pathogenic effects caused by particular CAstV infections may well correlate with the extent of virus replication in the intestine or in other internal organs such as the kidney, which is known to be infected by some CAstVs (Smyth et al., Citation2007). The development and application of a real-time RT-PCR test with the capacity to quantify virus load are underway to address these issues.
References
- Baxendale , W. and Mebatsion , T. 2004 . The isolation and characterisation of astroviruses from chickens . Avian Pathology , 33 : 364 – 370 .
- Day , J.M. , Spackman , E. and Pantin-Jackwood , M. 2007 . A multiplex RT-PCR test for the differential identification of turkey astrovirus type 1, turkey astrovirus type 2, chicken astrovirus, avian nephritis virus and avian rotavirus . Avian Diseases , 51 : 681 – 684 .
- Decaesstecker , M. , Charlier , G. and Meulemans , G. 1986 . Significance of parvoviruses, enterovirus-like viruses and reoviruses in the aetiology of the chicken malabsorption syndrome . Avian Pathology , 15 : 769 – 782 .
- de Wit , J.J. 2008 . Practical epidemiology of poultry disease and multifactorial conditions . In M. Pattison , P.F. McMullin , J.M. Bradbury & D.J. Alexander Poultry Diseases 6th edn 503 506 . London : Saunders Elsevier .
- Imada , T. , Yamaguchi , S. , Mase , M. , Tsukamoto , K. , Kubo , M. and Morooka , A . 2000 . Avian nephritis virus (ANV) as a new member of the family Astroviridae and construction of infectious ANV cDNA . Journal of Virology , 74 : 8487 – 8493 .
- Koci , M.D. , Seal , B.S. and Schultz-Cherry , S. 2000 . Development of an RT-PCR diagnostic test for an avian astrovirus . Journal of Virological Methods , 90 : 79 – 83 .
- McNeilly , F. , Connor , T.J. , Calvert , V.M. , Smyth , J.A. , Curran , W.L. Morley , A.J. 1994 . Studies on a new enterovirus-like virus isolated from chickens . Avian Pathology , 23 : 313 – 327 .
- McNulty , M.S. , Allan , G.M. , Connor , T.J. , McFerran , J.B. and McCracken , R.M. 1984 . An entero-like virus associated with the runting syndrome in broiler chickens . Avian Pathology , 13 : 429 – 439 .
- Madeley , C.R. and Cosgrove , B.P. 1975 . Letter: 28 nm particles in faeces in infantile gastroenteritis . Lancet , 2 : 451 – 452 .
- Méndez , E. Arias , C.F. 2007 . Astroviruses . In D.M. Knipe & P.M. Howley Fields Virology 5th edn 1 , 981 1000 Philadelphia , PA : Lippincott Williams & Wilkins .
- McNulty , M.S. , Connor , T.J. , McNeilly , F. and McNeilly , J.B. 1990 . Biological characterisation of avian enteroviruses and enterovirus-like viruses . Avian Pathology , 19 : 75 – 87 .
- Pantin-Jackwood , M.J. , Spackman , E. and Woolcock , P.R. 2006 . Molecular characterization and typing of chicken and turkey Astroviruses circulating in the United States: implications for diagnostics . Avian Diseases , 50 : 397 – 404 .
- Pantin-Jackwood , M.J. , Spackman , E. , Day , J.M. and Rives , D. 2007 . Periodic monitoring of commercial turkeys for enteric viruses indicates continuous presence of astrovirus and rotavirus on the farms . Avian Diseases , 51 : 674 – 680 .
- Reynolds , D.L. , Saif , Y.M. and Theil , K.W. 1987 . A survey of enteric viruses of turkey poults . Avian Diseases , 31 : 89 – 98 .
- Spackman , D. , Gough , R.E. , Collins , M.S. and Lanning , D. 1984 . Isolation of an enterovirus-like agent from the meconium of dead-in-shell chicken embryos . Veterinary Record , 114 : 216 – 218 .
- Smyth , J.A. , Connor , T.J. , McNeilly , F. , Moffet , D.A. , Calvert , V.M. and McNulty , M.S. 2007 . Studies on the pathogenicity of enterovirus-like viruses in chickens . Avian Pathology , 36 : 119 – 126 .
- Strain , E. , Kelley , L.A. , Schultz-Cherry , S. , Muse , S.V. and Koci , M.D. 2008 . Genomic analysis of closely related astroviruses . Journal of Virology , 82 : 5099 – 5103 .
- Tang , Y. , Ismail , M.M. and Saif , Y.M. 2004 . Development of antigen-capture enzyme-linked immunosorbent assay and RT-PCR for detection of turkey astroviruses . Avian Diseases , 49 : 182 – 188 .
- Tamura , K , Dudley , J. , Nei , M. Kumar , S. 2007 MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0 . Molecular Biology and Evolution , 24 , 1596 1599 . Retrieved November 2008, from http://www.megasoftware.net
- Todd , D. , Smyth , V.J. , Ball , N.W. , Donnelly , B.M. , Wylie , M. Knowles , N.J. 2009a . Characterisation of chicken enterovirus-like viruses, duck hepatitis virus (DHV) type 2 and DHV type 3 as astroviruses . Avian Pathology , 38 : 21 – 29 .
- Todd , D. , Wilkinson , D.S. , Jewhurst , H.L. , Wylie , M. , Gordon , A.W. and Adair , B.M. 2009b . A seroprevalence investigation of chicken astrovirus infections . Avian Pathology , 38 : 301 – 309 .