Abstract
Serovar and antimicrobial resistance data from the scanning surveillance of British turkey flocks for Salmonella between 1995 and 2006 were analysed and compared with prevalence data from other livestock and animal feed. A total of 2753 incidents of 57 different serovars were reported. The five most prevalent serovars were Salmonella Typhimurium (20.8%), Salmonella Newport (14.7%), Salmonella Derby (10.6%), Salmonella Indiana (8.3%) and Salmonella Agona (6.4%). S. Typhimurium reports peaked in the mid- to late 1990s; this occurred in parallel with the S. Typhimurium DT104 epidemic in other livestock species. S. Enteritidis reports peaked in mid- to late 1990s, followed by a considerable decrease after 2000, which was also noted in flocks of domestic fowl. S. Newport, Salmonella Montevideo, Salmonella Senftenberg and Salmonella Binza occurred in marked clusters, indicating that they were introduced into one or more flocks at a certain time (i.e. via contaminated feed or infected 1-day-old chicks). A proportion of 43.1% of the reported Salmonella isolates were resistant to at least one antimicrobial, while 17.7% were multi-resistant. No isolates were resistant to ciprofloxacin or to the third-generation cephalosporins ceftazidime and cefotaxime. Resistance to ampicillin, chloramphenicol, streptomycin, sulphonamide compounds and tetracycline was common, and it was mainly a characteristic of S. Typhimurium DT104 compared with S. Typhimurium non-DT104 and non-S. Typhimurium isolates (P<0.001). Resistance to nalidixic acid decreased from 16.9% in 1995 to 11.8% in 2006. Nalidixic acid resistance was most frequently found in Salonella Hadar (71.4%), S. Typhimurium DT104 (30.0%), S. Newport (17.9%) and S. Typhimurium non-DT104 (11.1%).
Introduction
Salmonellosis in humans is a major public health problem worldwide. In 2007 salmonellosis remained the second most commonly reported human zoonosis in the European Union (EU) in spite of a decrease in incidence over the past 4 years (European Food Safety Authority [EFSA], Citation2009a). The most commonly identified causative agent in the food-poisoning outbreaks in the UK in 2007 was Salmonella (EFSA, Citation2009b). Poultry-borne transmission is one route for human Salmonella infections. In 2007 around 8.0% of the fattening turkey flocks in the EU tested positive for Salmonella, while in 2005 to 2007 1.5% or less of the turkey production flocks in the EU were positive for Salmonella Enteritidis or Salmonella Typhimurium (with the exception of a single country) (EFSA, Citation2009a). Salmonella can be introduced into a turkey site potentially at all stages in the production pyramid and it can be transmitted vertically and horizontally. Turkey breeding flocks and hatcheries are critical sources of Salmonella, and it has been reported that turkey flocks may remain infected with this organism throughout the growing period (Cox et al., Citation2000). Feed has also been reported as a common source of Salmonella in turkey flocks (Hafez & Jodas, Citation2000).
Many Salmonella serovars have been isolated from turkey flocks; some may be predominant for many years in a certain region or country and then disappear to be replaced by another serovar (Hafez & Jodas, Citation2000). Poppe et al. (Citation1995) reported 52 serovars among 2690 isolates recovered from turkeys in Canada, Schroeter et al. (Citation1998) found 15 serovars among 658 isolates from turkey flocks in Germany, while Pedersen et al. (Citation2002) reported 24 different serovars among 234 isolates from turkey flocks sampled before slaughter in Denmark. The definitive phage type DT104 has been reported as being dominant among S. Typhimurium in turkey flocks in Germany (Schroeter et al., Citation1998) and Britain (Davies et al., Citation1999), but not in Denmark (Pedersen et al., Citation2002). Salmonella isolates from turkeys have been reported as having high levels of antimicrobial resistance (Zhao et al., Citation2007) and multi-resistance (Schroeter et al., Citation1998), and as being more frequently resistant than isolates from other livestock species (Schroeter et al., Citation1998; Poppe et al., Citation2001). Therefore Salmonella in turkeys and turkey meat should be regarded as an issue of great public health significance. The increasing occurrence of quinolone resistance in Salmonella isolates from food animal sources has been reported as a matter of concern (Griggs et al., Citation1994).
The UK is among the top countries worldwide for both exports and imports of turkey meat (Anonymous, Citation2008a). Between 1996 and 2005 the UK consumption of turkey meat remained fairly static, with approximately 4.0 to 5.1 kg/person per annum, even though the number of turkeys raised for human consumption on UK farms decreased from approximately 40.2 million in 1997 to 17.1 million in 2006 (Anonymous, Citation2008a). Recent census data show that there are around 11.6 million turkeys in 2423 flocks in Great Britain. Breeding and rearing sites and hatcheries may be subscribed to the Poultry Health Scheme. This is a trade facilitation scheme established to implement a system of approval for establishments officially recognized as meeting legal requirements covering the animal health conditions for trade in poultry and hatching eggs. Breeding and rearing sites and hatcheries subscribed to the Poultry Health Scheme are required to collect samples from the birds for Salmonella testing soon after they enter the laying period (Department of Environment, Food and Rural Affairs, Citation2008). Fattening flocks belonging to major turkey producing companies are voluntarily monitored for Salmonella during the rearing and during the finishing period.
The present paper reports results from the scanning surveillance of turkey flocks for Salmonella in Great Britain and puts these into context of expected prevalences and international data for major turkey producing nations. Salmonella serotype/phage type/resistance trends are compared with trends in feed (as a potential source) and in pigs (as a parallel source of S. Typhimurium).
Materials and Methods
Statutory reporting of Salmonella isolates
In Great Britain, Salmonella isolated from animals, their environment or animal feeding stuffs is reported to the Competent Authority under the Zoonoses Order 1989. In practice these reports are made to government veterinarians at the Veterinary Laboratories Agency (VLA) and the Salmonella isolates are submitted to the VLA Reference Laboratory for further identification, which includes serotyping, phage typing and analysis for resistance to a panel of 16 antimicrobials. Scanning surveillance data related to Salmonella incidents reported from turkey flocks in Great Britain from 1995 to 2006 were analysed retrospectively as part of our study.
A Salmonella incident report is defined as one in which Salmonella has been isolated from animals on a premises or from environmental samples from the premises and there have been no previous isolations of the same serotype or phage type in the same epidemiological group within the past 30 days. The concept of an “incident” is used in order to provide more representative data on the occurrence of Salmonella in the animal population, as the intensity of sampling (i.e. number of samples collected from a particular epidemiological group) can vary. The designation of Salmonella incidents therefore assists in minimizing bias, which arises as a result of differences in the intensity of sampling, and the 30-day period has been adopted to standardize procedures as detailed data on individual holdings are often not available.
Data sources
The data sources analysed in this paper included the diagnostic investigation of animal disease by veterinarians as well as the voluntary monitoring for Salmonella undertaken by the turkey industry. The interpretation of the latter is complicated by the variability between companies and changes in the level and sensitivity of monitoring over time. Furthermore, data from the voluntary monitoring of turkey breeding flocks under the Poultry Breeding Flocks and Hatcheries Order 1993 during the rearing and laying period, under the Poultry Health Scheme soon after the start of lay and data from zoonotic investigations at farms under the Zoonoses Order 1989 were included in the analysis. Data analysed in this paper originated from sampling of breeding and meat production flocks conducted on farms or hatcheries. Data of non-Great Britain origin (i.e. imported eggs or 1-day-old chicks sampled in Great Britain) were excluded from the analysis.
Serotyping and phage typing methods
Salmonella isolates were biochemically confirmed and serotyped by micro titres and tube and slide agglutination tests, respectively. Serovars were derived by reference to the Kauffmann–White Scheme (Popoff, Citation2001). The serotyping methods used were the same throughout the study period. S. Enteritidis, Salmonella Hadar, S. Typhimurium and Salmonella Virchow were phage typed according to the Health Protection Agency phage typing schemes (Callow, Citation1959; Anderson et al., Citation1977; De Sa et al., Citation1980; Chambers et al., Citation1987; Ward et al., Citation1987).
Antimicrobial sensitivity testing
Salmonella isolates received for serotyping were tested for their in vitro sensitivity against a panel of 16 antimicrobials. The historical veterinary breakpoint of 13 mm was used across the board of antimicrobials (Chris J. Teale, personal communication); that is, isolates were considered resistant when the growth inhibition zone around the disk had a diameter of ≤13 mm and the cultures were classified as either resistant or sensitive. All tests were performed using a disk diffusion technique using Oxoid “Isosensitest” agar (Sensitest agar was used prior to 2000; Chris J. Teale, personal communication) and antimicrobial-containing disks (Wray et al., Citation1991; Jones et al., Citation2002). The disks contained the following antimicrobial agents: amikacin (30 µg), amoxycillin/clavulanic acid (30 µg), ampicillin (10 µg), apramycin (15 µg), cefoperazone (30 µg (replaced with 30 µg cefotaxime disc in 2004)), cefuroxime (30 µg (replaced with 30 µg ceftazidime disc in 2001)), chloramphenicol (10 µg), tetracycline (10 µg), colistin (25 µg (replaced with 1 µg ciprofloxacin disc in 2004)), furazolidone (15 µg), gentamicin (20 µg (10 µg from 1998)), nalidixic acid (30 µg), neomycin (10 µg), streptomycin (25 µg), sulphamethoxazole/trimethoprim (25 µg), and sulphonamide compounds (500 µg (300 µg from 1998)). Since 1996 only the first isolate from each incident has been tested for its antimicrobial susceptibility. This is done because some organizations (particularly poultry companies) perform extensive monitoring for Salmonella, and this can skew the overall susceptibility data reflecting the intensity of sampling procedures.
Data analysis
Data were retrieved from the VLA Farmfile Salmonella database and summarized using Business Objects version 6. Univariate analysis was conducted with the chi-squared test at P=0.05 using Microsoft Excel Statistics 2003 for Windows to detect associations between Salmonella serovars and antimicrobial resistance.
Results
Most prevalent Salmonella serovars
A total of 2753 incidents of 57 different serovars were reported. The five most prevalent serovars, which represented 60.8% of the total number of reports, were S. Typhimurium, Salmonella Newport, Salmonella Derby, Salmonella Indiana and Salmonella Agona (20.8%, 14.7%, 10.6%, 8.3% and 6.4% of all incidents, respectively; ). Between 1995 and 2006 the proportion of S. Typhimurium and S. Newport reports decreased from 28.1% and 28.8% to 21.7% and 7.8%, respectively, while S. Derby and S. Agona reports increased from 5.0% and 2.0% to 15.0% and 5.0%, respectively. The proportion of S. Indiana fluctuated considerably over the years without any clear trend being evident. Thirty-five per cent of S. Newport incidents were reported in 1995 to 1996. The reporting frequency of S. Derby increased since 1999, and it has recently become one of the most common serovars in British turkeys. S. Agona was also reported frequently from turkeys after 1999. Other prevalent serovars were Salmonella Montevideo and Salmonella Kottbus (each accounted for 5.6% of the reported incidents). Clustering was observed in S. Montevideo between 2000 and 2003 when 79.3% of the incidents were reported, while the proportion of reported incidents of S. Kottbus increased from 2.9% in 1999 to 13.9% in 2006. Salmonella Kedougou was one of the most frequently reported serovars between 2004 and 2006 (proportion of reported incidents ranged from 7.0% to 11.1%) but it was rarely reported in earlier years. Reports of Salmonella Senftenberg peaked in 1998 (9.2%). Salmonella Binza accounted for 2.1% of all incidents, but it was mostly prevalent in the first half of the study period. S. Hadar and S. Virchow each accounted for 1.6% of the reported incidents, and the latter was mainly reported since 2003.
Table 1. Frequency distribution (number) of most common Salmonella serovars in turkeys in 2006 in comparison with previous years
Salmonella serovars reported consistently all years included S. Agona, S. Derby, S. Indiana, S. Kedougou, S. Newport and S. Typhimurium. Thirteen serovars were reported only once. These were Salmonella Alachua (in 1997), Salmonella Drypool (in 1995), Salmonella Hato (in 2001), Salmonella Kokomlemle (in 1995), Salmonella Liverpool (in 1996), Salmonella Livingstone (in 1997), Salmonella Manhattan (in 2003), Salmonella Muenster (in 1999), Salmonella Stanley (in 2002), Salmonella Stourbridge (in 2006), Salmonella Teddington (in 2001), Salmonella Thomasville (in 1997) and Salmonella Thompson (in 1996).
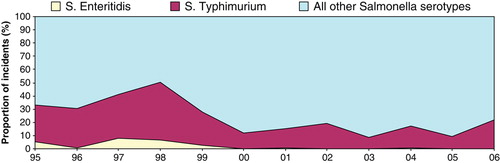
Trends in S. Enteritidis and S. Typhimurium over time
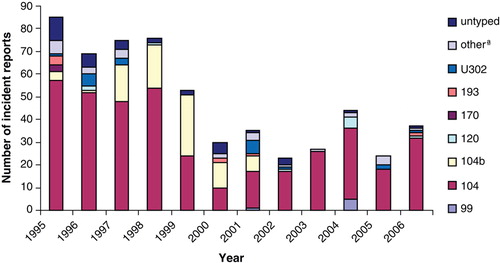
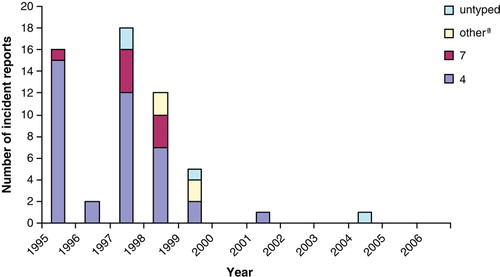
S. Typhimurium was the predominant serovar in turkeys but S. Enteritidis reports were rare (55 in total). There was clustering in reports of both serovars between 1995 and 1999, when 61.6% of S. Typhimurium and 96.4% of S. Enteritidis were reported (). Various definitive types (DTs) of S. Typhimurium, including DT99, DT120, DT170, DT193 and U302, were reported, but DT104 and DT104b were the most prevalent (). The S. Enteritidis isolates belonged mainly to phage type (PT) 4, while PT1, PT6, PT7, PT9b and PT36 were reported occasionally ().
Antimicrobial sensitivity trends
A total 43.1% of the Salmonella isolates that were tested for their antimicrobial resistance were resistant to at least one antimicrobial, and 17.7% were multi-resistant (resistant to more than three unrelated antimicrobials) (). The highest resistance was recorded to sulphonamide compounds (33.1%), tetracycline (29.7%), streptomycin (23.6%), ampicillin (20.5%), nalidixic acid (17.9%) and chloramphenicol (14.2%). Isolates belonging to 52 different serovars, and most frequently Salmonella Mbandaka, S. Kottbus and S. Derby, were fully sensitive to all the antimicrobials tested against (data not presented). Multi-resistance was recorded in isolates belonging to 17 different serovars, and it was most common in S. Typhimurium, S. Newport and S. Indiana (data not presented). There were no isolates resistant to amikacin, amoxicillin/clavulanic acid, gentamicin, ceftazidime, cefotaxime or to ciprofloxacin. S. Typhimurium, and especially DT104 compared with non-DT104 isolates, were more likely to be resistant to any unrelated antimicrobial (P<0.001), and more specifically to streptomycin (P<0.001), sulphonamide compounds (P<0.001), tetracycline (P<0.001), ampicillin (P<0.001), chloramphenicol (P<0.001) and nalidixic acid (P<0.001) compared with non-S. Typhimurium isolates. Non-S. Typhimurium isolates were more likely to be resistant to sulphamethoxazole/trimethoprim (P<0.001) compared with S. Typhimurium isolates. Resistance of S. Typhimurium DT104, S. Typhimurium non-DT104 and non-S. Typhimurium isolates is presented in and and Supplementary . Various serovars were resistant to nalidixic acid (Supplementary ) but most frequently S. Hadar (71.4%), S. Typhimurium DT104 (30.0%), S. Newport (17.9%) and S. Typhimurium non-DT104 (11.1%). A total 17.9% of all Salmonella isolates tested in the study period were resistant to nalidixic acid, but resistance to this antimicrobial decreased from 16.9% in 1995 to 11.8% in 2006 (). Resistance to nalidixic acid was significantly higher among isolates of S. Typhimurium DT104 as compared with those of S. Typhimurium non-DT104 (P<0.001), S. Newport (P<0.001), S. Kottbus (P<0.001), S. Indiana (P<0.001), S. Kedougou (P<0.001), S. Montevideo (P<0.001) and S. Senftenberg (P<0.001), but significantly lower compared with S. Hadar (P<0.001). S. Typhimurium non-DT104 isolates were more frequently resistant to nalidixic acid compared with those of S. Kedougou (P=0.002), S. Kottbus (P<0.001), S. Indiana (P<0.001) and S. Montevideo (P=0.001), but less frequently resistant compared with S. Newport (P=0.021) and S. Hadar (P<0.001). S. Newport isolates were more frequently resistant to nalidixic acid compared with isolates of S. Indiana (P<0.001), S. Kottbus (P<0.001), S. Montevideo (P<0.001), S. Senftenberg (P=0.007) and S. Kedougou (P<0.001), but less frequently resistant compared with S. Hadar (P<0.001).
Table 2. Salmonella serovars from turkeys: antimicrobial sensitivity monitoring in 1995–2006 (N= number of cultures tested, %R = % of isolates resistant to at least 1 antimicrobial, %R3 = % of isolates resistant to more than 3 antimicrobials)
Table 3. S. Typhimurium DT104a from turkeys: antimicrobial sensitivity monitoring in 1995–2006
Table 4. S. Typhimurium non-DT104a from turkeys: antimicrobial sensitivity monitoring in 1995-2006
Discussion
The range of Salmonella serovars reported from British turkey flocks suggests a variety of sources, such as contaminated feed, infected breeding flocks, persistent infection in hatcheries, or contaminated rearing sites. The fact that various serovars were reported only once before disappearing and the clustering in reports of S. Newport, S. Montevideo, S. Senftenberg and S. Binza suggest that they were introduced into one or more flocks at a certain time (i.e. via contaminated feed or infected 1-day-old chicks) and they were subsequently controlled or eradicated. This has been reported previously (Pedersen et al., Citation2002).
S. Derby and S. Agona, which were among the most prevalent serovars in British turkey flocks in our study, have also been reported as common strains in Danish turkeys (Pedersen et al., Citation2002), and the latter has also been reported as the most prevalent in commercial turkey flocks in Canada (Irwin et al., Citation1994). Comparisons with other studies should be made with caution due to differences in monitoring schemes and/or study design, laboratory methodologies applied and reporting differences. It is clear from these reports as well as the recent EU baseline survey for Salmonella in turkeys (EFSA, Citation2008) that certain serovars are widely distributed in turkeys, which is likely to reflect international trade in breeding turkeys, 1-day-old poults and contaminated feed ingredients.
S. Typhimurium was the predominant serovar in turkeys in Great Britain throughout the study period with a peak in reports in the mid to late 1990s. This occurred in parallel with the S. Typhimurium DT104 epidemic in other livestock species, such as cattle and pigs (Anonymous, Citation2007). This serovar was reported routinely from primary breeders in Great Britain in 1995 to 1997 (data not presented). The subsequent fluctuation of reports of S. Typhimurium in turkeys could possibly be due to influence of company sampling and reporting programme changes associated with changes in ownership. In 2006 S. Typhimurium was the most frequently reported serovar from turkeys in Great Britain under routine surveillance (reported at a frequency of 21.7%), but it was second in rank in turkey holdings in the UK found positive for Salmonella in the EU survey carried out in 2006 to 2007 (EFSA, Citation2008). The difference between the distribution of serovars reported under routine surveillance in Great Britain and that from the EU survey could be due to the fact that scanning surveillance data originate from the routine monitoring and from the investigation of clinical disease undertaken by major turkey companies for Salmonella in Great Britain. It could also be skewed by enhanced monitoring undertaken by the poultry industry for Salmonella control purposes, while the mandatory EU survey data resulted from a random sample of turkey flocks in the country and included data from a large number of seasonal producers who do not routinely test their flocks for Salmonella. However, more recent surveillance data support a further decrease in the reporting frequency of S. Typhimurium from British turkey flocks in 2007 in comparison with earlier years (Anonymous, Citation2008b). Other countries that have not reported S. Typhimurium as a common turkey serovar include Denmark (Pedersen et al., Citation2002) and the US (Santos et al., Citation2007). S. Typhimurium control programs in the US, which is a major turkey-producing nation, had aimed historically at reducing the infection of S. Typhimurium in turkey flocks (Kumar et al., Citation1971). Various definitive types of S. Typhimurium were reported in our study, but DT104 was predominant. The main source of DT104 in turkeys is thought to have been resident infection on some rearing and fattening units following historic introduction via spread from cattle, breeding stock or contaminated feed, but this situation has been addressed more successfully in recent years. In a study conducted in Denmark to investigate the Salmonella prevalence in turkey flocks between 1995 and 2000, none of the six S. Typhimurium isolates belonged to phage type DT104 (Pedersen et al., Citation2002). However, phage typing may not always be done routinely in countries that find S. Typhimurium in turkeys. DT104 was the predominant S. Typhimurium definitive type in pigs in Great Britain until 2000, and remains so in cattle (Anonymous, Citation2007). However, the other common S. Typhimurium DTs in pigs, such as DT208, and especially U302 and U288 in recent years (data not presented), were rarely reported from turkeys. It is also interesting to note that the S. Typhimurium DT56, DT40 and DT41, which are commonly found in wild birds (VLA, Citation2008), were rarely reported from turkeys in Great Britain despite concern about contact of wild with domestic birds in free-range and semi-open housing. These DTs are not very infectious for domestic poultry, but are host-adapted in wild birds (Hughes et al., Citation2008).
The peak in S. Enteritidis in British turkey flocks in the mid to late 1990s, and the subsequent decrease, was also seen in flocks of domestic fowl (Anonymous, Citation2007). These data are also in agreement with the 2006 to 2007 EU Salmonella in turkeys survey UK results: S. Enteritidis was not isolated from any turkey breeding or fattening holding in the UK. The predominant phage type of S. Enteritidis in turkeys was PT4, and PT7, PT1 and PT6 were also occasionally reported. The fact that these are mainly chicken-related PTs could possibly indicate indirect spread of infection from chickens via contaminated waste, wildlife, bedding, and so on. This is supported by the trends in S. Enteritidis prevalence figures from chickens, which demonstrate that S. Enteritidis was only prevalent in turkeys when the prevalence in chickens was high (Anonymous, Citation2007).
Even though from 1996 and according to the guidelines for susceptibility testing only one isolate from each incident should have been selected for testing, the data presented in and suggest that this may not always have happened; for example, if this had always happened, 2454 Salmonella cultures should have been selected for susceptibility testing from 1996 to 2006, but in fact 4663 cultures were tested. As a result, isolates may have been selected for testing in a non-representative way and this may have biased the validity of the data presented in this paper. A high proportion (43.1%) of Salmonella isolates from British turkey flocks was found to be resistant to at least one antimicrobial while 17.7% of the isolates were multi-resistant. These figures are lower compared with those published for Salmonella isolates recovered from turkey diagnostic samples in the US between 2002 and 2003 (Zhao et al., Citation2007), but higher compared with those reported for Danish turkey flocks (Pedersen et al., Citation2002). No isolates in our study were found to be resistant to ciprofloxacin, which was in agreement with data reported from Canada (Poppe et al., Citation1995), or to the third-generation cephalosporins ceftazidime and cefotaxime, or to cefuroxime. This observation is important, as these compounds are antimicrobials of choice in human patients for treatment of salmonellosis. Ceftiofur resistance has been reported from Salmonella isolated from turkeys in the US. (Frye & Fedorka-Cray, Citation2007). Resistance to ampicillin, chloramphenicol, streptomycin, sulphonamide compounds and tetracycline was common and it was mainly characteristic of S. Typhimurium DT104 isolates. This may be associated with tetracycline and ampicillin usage as antimicrobials (Nde & Logue, Citation2007) but the role of clonal spread is likely to be predominant. The pentavalent resistant S. Typhimurium DT104 strains are still prevalent in turkeys, unlike pigs in which there seems to have been an increase in resistance to the four antimicrobials ampicillin, streptomycin, sulphonamide compounds and tetracycline but not to chloramphenicol in currently circulating strains (Anonymous, Citation2007).
Data indicate that antimicrobial resistance levels (and especially resistance to ampicillin, chloramphenicol, streptomycin, sulphonamide compounds, tetracycline and nalidixic acid) for S. Typhimurium isolates in 2000 were much lower compared with other years. This was unlikely to have been due to a change in laboratory methodologies as, if this had been the case, it would have affected the results of more than one species and it would have had an ongoing effect from that point onwards. Both scenarios were checked and neither was found to b e true. It is more likely that this deviation reflects either changes in the sources of the Salmonella isolates (e.g. a large turkey company either ceasing or starting to submit isolates) or extensive monitoring occurring on a set of related premises that were affected by the same strain. For these reasons, the data in 2000 should not be considered directly comparable with other years.
The decrease in the proportion of Salmonella isolates from turkeys in Great Britain tested resistant to nalidixic acid from 16.9% in 1995 to 11.8% in 2006 probably reflects prudent usage initiatives, but this could not be verified against available antimicrobial usage data, which do not differentiate use by different species. From 1995 to 1998 fluoroquinolone sales in the UK remained at around 1 tonne of active ingredient since they were first authorized in 1993 (VMD, Citation2008a), whereas in each of the nine reporting years from 1998 to 2006 between 1 and 2 tonnes of fluoroquinolones were sold (VMD, Citation2008b). Even though resistance of Salmonella turkey isolates to nalidixic acid was still relatively common (11.8%) in 2006, it was not at that time much higher than chickens (10.6%) (Anonymous, Citation2007). The proportion of Salmonella reported from other livestock species in 2006 that were resistant to nalidixic acid (1.1% in cattle and 2.9% in pigs) was much lower than poultry (Anonymous, Citation2007). Although various serovars from turkeys, and especially S. Hadar, S. Typhimurium and S. Newport, were resistant to nalidixic acid in the study period, there are certain serovars, such as S. Enteritidis, S. Virchow, and S. Derby, which did not exhibit resistance to this antimicrobial even though they were possibly subjected to the same selective pressure.
In spite of the mandatory nature of the reporting of Salmonella isolates to the VLA, it is unknown how Salmonella monitoring practices in the turkey industry may have changed over time and this could have affected the representativeness of the data presented in this paper. National control programmes for Salmonella in fattening and breeding turkeys are planned to be implemented in January 2010 to comply with Regulation (EC) No 2160/2003 and Regulation (EC) No 584/2008. As a result, monitoring of turkey flocks for Salmonella in the UK as in all EU countries will become mandatory and it is expected that the current surveillance system will be standardized and improved. More importantly, control systems will be put in place for the reduction of the maximum percentage of British turkey flocks remaining positive for S. Enteritidis and S. Typhimurium to no more than 1% by the end of 2012. According to results of the EU Salmonella in turkeys survey (EFSA, Citation2008), the Salmonella prevalence in UK turkey breeding flocks was found within this level (0.5%, 95% confidence interval = 0.1 to 3.2%), but this will be a substantial target to achieve for turkey fattening flocks as the baseline survey estimated Salmonella prevalence in turkey fatteners in UK at 4.6% (95% confidence interval = 2.2 to 9.0%).
Supplementary Table 1. Salmonella serovars other than S. Typhimurium a from turkeys: antimicrobial sensitivity monitoring in 1995–2006
Supplementary Table 2. Percentage of Salmonella serovars from turkeys resistant to nalidixic acid in 1995-2006 (number of isolates tested is presented in parenthesis)
Acknowledgements
The present work was funded by the UK Department of Environment, Food and Rural Affairs. The authors would like to thank Ms Sue Kidd and Mr Robert Smeatham (Centre for Epidemiology and Risk Analysis, VLA) for assistance in preparation of the data.
References
- Anderson , E.S. , Ward , L.R. , De Saxe , M.J. and De Sa , J.D.H. 1977 . Bacteriophage—typing designations of Salmonella typhimurium . Journal of Hygiene , 78 : 297 – 300 .
- Anonymous 2007 Salmonella in Livestock Production 2006 . DEFRA , London , , UK .
- Anonymous 2008a ADAS, Defra Funded Project OZ0328—A Status Report on the UK Turkey Industry . VLA , Weybridge , , UK .
- Anonymous 2008b Salmonella in Livestock Production 2007 . DEFRA , London , , UK .
- Callow , B.R. 1959 . A new phage-typing scheme for Salmonella typhimurium . Journal of Hygiene , 57 : 346 – 359 .
- Chambers , R. , McAdam , P. , De Sa , J.D.H. , Ward , L.R. and Rowe , B. 1987 . A phage typing scheme for Salmonella virchow . FEMS Microbiology Letters , 40 : 155 – 157 .
- Cox , N.A. , Stern , N.J. , Craven , S.E. , Berrang , M.E. and Musgrove , M.T. 2000 . Prevalence of Campylobacter and Salmonella in the cecal droppings of turkeys during production . Journal of Applied Poultry Research , 9 : 542 – 545 .
- Davies , R.H. , Teale , C.J. , Wray , C. , McLaren , I.M. , Jones , Y.E. , Chapell , S. and Kidd , S. 1999 . Nalidixic acid resistance in salmonellae isolated from turkeys and other livestock in Great Britain . Veterinary Record , 144 : 320 – 322 .
- Department of Environment, Food and Rural Affairs 2008 Poultry Health Scheme Members’ Handbook Available online at http://www.defra.gov.uk/animalh/int-trde/eu/animals/pdf/poultry_health_eng.pdf (accessed 12 June 2008) .
- De Sa , J.D.H. , Ward , L.R. and Rowe , B. 1980 . A scheme for phage typing of Salmonella hadar . FEMS Microbiology Letter , 9 : 175 – 177 .
- European Food Safety Authority 2008 Report of the Task Force on Zoonoses Data Collection on the Analysis of the baseline survey on the prevalence of Salmonella in turkey flocks, in the EU, 2006–2007 . The EFSA Journal , 134 , 1 – 91 . Available online at http://www.efsa.europa.eu/EFSA/efsa_locale-1178620753812_1178706574172.htm (accessed 15 May 2008) .
- European Food Safety Authority 2009a The Community Summary Report on Trends and Sources of Zoonoses, Zoonotic Agents, Antimicrobial Resistance and Foodborne Outbreaks in the European Union in 2007 . Available online at : http://www.efsa.europa.eu/cs/BlobServer/Report/2007_Zoonoses_Community_Summary_Report,0.pdf?ssbinary=true (accessed 3 February 2009) .
- European Food Safety Authority 2009b United Kingdom—Trends and Sources of Zoonoses and Zoonotic Agents in Humans, Foodstuffs, Animals and Feedingstuffs in 2007 . Available online at : http://www.efsa.europa.eu/cs/BlobServer/DocumentSet/United_Kingdom_2007.pdf?ssbinary=true (accessed 3 February 2009) .
- Frye , J.G. and Fedorka-Cray , P.J. 2007 . Prevalence, distribution and characterisation of ceftiofur resistance in Salmonella enterica isolated from animals in the USA from 1999 to 2003 . International Journal of Antimicrobial Agents , 30 : 134 – 142 .
- Griggs , D.J. , Hall , M.C. , Jin , Y.F. and Piddock , L.J. 1994 . Quinolone resistance in veterinary isolates of salmonella . Journal of Antimicrobial Chemotherapy , 33 : 1173 – 1189 .
- Hafez , H.M. and Jodas , S. 2000 . “ Salmonella infections in turkeys ” . In Salmonella in Domestic Animals , 1st edn , Edited by: Wray , C. and Wray , A. 133 – 155 . Wallingford : CABI Publishing .
- Hughes , L.A. , Shopland , S. , Wigley , P. , Bradon , H. , Leatherbarrow , A.H. Williams , N.J. 2008 . Characterisation of Salmonella enterica serotype Typhimurium isolates from wild birds in northern England from 2005–2006 . BMC Veterinary Research , 4 : 4
- Irwin , R.J. , Poppe , C. , Messier , S. , Finley , G.G. and Oggel , J. 1994 . A national survey to estimate the prevalence of Salmonella species among Canadian registered commercial turkey flocks . Canadian Journal of Veterinary Research , 58 : 263 – 267 .
- Jones , Y.E. , Chappell , S. , McLaren , I.M. , Davies , R.H. and Wray , C. 2002 . Antimicrobial resistance in Salmonella isolated from animals and their environment in England and Wales from 1988 to 1999 . The Veterinary Record , 150 : 649 – 654 .
- Kumar , M.C. , York , M.D. , McDowell , J.R. and Pomeroy , B.S. 1971 . Dynamics of Salmonella infection in fryer roaster turkeys . Journal of Applied Poultry Research , 15 : 221 – 232 .
- Nde , C.W. and Logue , C.M. 2007 . Characterization of antimicrobial susceptibility and virulence genes of Salmonella serovars collected at a commercial turkey processing plant . Journal of Applied Microbiology , 104 : 215 – 223 .
- Pedersen , K. , Hansen , H.C. , Jorgensen , J.C. and Borck , B. 2002 . Serovars of Salmonella isolated from Danish turkeys between 1995 and 2000 and their antimicrobial resistance . The Veterinary Record , 150 : 471 – 474 .
- Popoff , M.Y. 2001 . “ WHO Collaborating Centre for Reference and Research on Salmonella ” . In Antigenic formulas of the Salmonella serovars , Paris : Institute Pasteur .
- Poppe , C. , Kolar , J.J. , Demczuk , W.H.B. and Harris , J.E. 1995 . Drug-resistance and biochemical characteristics of Salmonella from turkeys . Canadian Journal of Veterinary Research , 59 : 241 – 248 .
- Poppe , C. , Ayroud , M. , Ollis , G. , Chirino-Trejo , M. , Smart , N. Quessy , S. 2001 . Trends in antimicrobial resistance of Salmonella isolated from animals, foods of animal origin, and the environment of animal production in Canada, 1994–1997 . Microbial Drug Resistance(Mechanisms Epidemiology and Disease , 7 : 197 – 212 .
- Santos , F.B. , DeSouza , D.H. , Jaykus , L. , Ferket , P.R. and Sheldon , B.W. 2007 . Genotypes, serotypes, and antibiotic resistance profiles of Salmonella isolated from commercial North Carolina turkey farms . Journal of Food Protection , 70 : 1328 – 1333 .
- Schroeter , A. , Rabsch , W. & Helmuth , R. 1998 The current situation on Salmonella in turkeys . In Proceedings of the 1 st International Symposium on Turkey Diseases 170 – 177 Berlin , Germany
- VLA 2008 Wildlife Diseases in the UK Reported in the Year 2006—Report to Defra and to OIE . Available online at http://www.defra.gov.uk/corporate/vla/science/documents/science-end-oie06.pdf (accessed 30 March 2008) .
- VMD 2008a Sales of Antimicrobial Products used as Veterinary Medicines, Growth Promoters and Coccidiostats in the UK from 1993–2000 . Available online at http://www.vmd.gov.uk/Publications/Antibiotic/salesanti9398.pdf (accessed 13 February 2008) .
- VMD 2008b Sales of Antimicrobial Products Authorized for Use as Veterinary Medicines, Antiprotozoals, Antifungals, Growth Promoters and Coccidiostats in the UK in 2006 . Available online at http://www.vmd.gov.uk/publications/antibiotic/salesanti06.pdf (accessed 13 February 2008) .
- Ward , L.R. , De Sa , J.D.H. and Rowe , B. 1987 . A phage-typing scheme for Salmonella enteritidis . Epidemiology and Infection , 99 : 291 – 294 .
- Wray , C. , Beedell , Y.E. and Mclaren , I.M. 1991 . A survey of antimicrobial resistance in salmonellae isolated from animals in England and Wales during 1984–1987 . British Veterinary Journal , 147 : 356 – 369 .
- Zhao , S. , McDermott , P.F. , White , D.G. , Qaiyumi , S. , Friedman , S. Abbott , J. 2007 . Characterization of multidrug resistant Salmonella recovered from diseased animals . Veterinary Microbiology , 123 : 122 – 132 .