Abstract
In this study, we report the development and validation of a duplex real-time polymerase chain reaction (PCR) assay with an internal control using TaqMan-labelled probes for the detection of Mycoplasma gallisepticum and Mycoplasma synoviae (duplex MGMS PCR). The MGMS PCR was highly specific with a sensitivity of 7 and 1 colony-forming units/ml for M. gallisepticum and M. synoviae, respectively, using dilution of pure culture that corresponds to 34 and 29 DNA copies per reaction. Validation of the assay was completed with 260 and 27 pooled samples (tracheal swabs) from commercial chickens and turkeys, respectively, with potential M. gallisepticum and M. synoviae involvement and 42 samples (palatine cleft swabs) from backyard geese and ducks. Using isolation as the gold standard, the MGMS PCR was more sensitive than isolation and the analytical sensitivity was 0.944 and 0.958 for M. gallisepticum and M. synoviae, respectively. In comparison with a gapA-based assay (gapA PCR) and a 16S rRNA-based assay (16S PCR) for M. gallisepticum and M. synoviae, respectively, the results agreed for 94.5% and 96.6%, respectively. The use of the internal control allowed monitoring of proper extraction and inhibition of amplification that was detected in 12 samples. The duplex MGMS PCR was shown to be superior to the presently reported real-time PCR assays in terms of combination of sensitivity, specificity and capacity of detection of more than one target in a single tube. In conclusion, the duplex MGMS PCR was highly specific, sensitive, and reproducible and could be used on clinical samples from commercial chickens, turkeys and backyard poultry including ducks and geese.
Introduction
Mycoplasma gallisepticum and Mycoplasma synoviae are major avian respiratory pathogens (Kleven, Citation1997; Ley & Yoder, Citation1997). M. gallisepticum causes acute and chronic respiratory diseases accompanied by catarrhal inflammation of respiratory tract mucosa (Ley, Citation2003). M. synoviae causes synovitis and airsacculitis (Bradbury, Citation2005). Reduced egg production and downgrading of carcass quality occur in infected poultry and can result in considerable economic losses. M. gallisepticum and M. synoviae can infect the embryo and subsequently produce infected progeny (Levisohn & Kleven, Citation2000). Concurrent M. synoviae infection with a secondary pathogen, such as Escherichia coli, can raise morbidity rates of infected birds (Raviv et al., Citation2007). The success of control programmes depends on accurate and timely diagnosis of infected flocks. Therefore, a diagnostic assay with high sensitivity, high specificity, and a fast detection time is required for monitoring M. gallisepticum and M. synoviae in poultry flocks including ducks and geese (Buntz et al., Citation1986; Benčina et al., Citation1988; Cookson & Shivaprasad, Citation1994). The gold standard for mycoplasma diagnosis is the isolation of mycoplasmas. Isolation and direct or indirect fluorescent antibody assays using specific polyclonal antisera are performed to differentiate M. gallisepticum colonies from colonies of other saprophytic mycoplasma species present in the upper respiratory tracts of normal birds (Kleven, Citation1998). Although the target mycoplasmas may be overgrown by saprophytic mycoplasmas, other conventional bacteria may impede isolation. Culturing of M. gallisepticum may take up to 2 weeks or more. Mycoplasma detection via serological methods may be performed in a short period of time—rapid plate agglutination test, haemagglutination inhibition, and enzyme-linked immunosorbent assay (ELISA) (Kleven, Citation1998). However, it has been shown that serological assays have drawbacks. Seroconversion lags behind infection. A minimum of 1 week after infection is required before antibodies can be detected by rapid plate agglutination (RPA), and 3 weeks are required to detect haemagglutination inhibition titres (Kleven, Citation1975; Georgiades et al., Citation2001). Furthermore, different groups have addressed the non-specific reactions caused by cross-reactions with other pathogenic mycoplasmas and the relatively low sensitivity of some serological assays (Avakian & Kleven, Citation1990; Kleven et al., Citation1996; Kempf et al., Citation1997).
An alternative to conventional culture and serology is the use of specific DNA detection methods. M. gallisepticum or M. synoviae may be detected by hybridization with DNA probes (Khan & Kleven, Citation1993), but now it is much more common to use the PCR to amplify specific portions of DNA in the test material (Nascimento et al., Citation1991; Marois et al., Citation2002). It is important to note that PCR assays based on the mgc2 gene of M. gallisepticum (Garcia et al., Citation2005) or the vlhA gene of M. synoviae (Hong et al., Citation2004) are becoming more widely used for PCRs for M. gallisepticum and M. synoviae. These genes have been reported suitable for mycoplasma detection (Garcia et al., Citation2005; Hammond et al., Citation2009). More recently, conventional PCR methods have been described for simultaneous detection of M. gallisepticum and M. synoviae (Garcia et al., Citation1995; Wang et al., Citation1997; Pang et al., Citation2002; Mardassi et al., Citation2005). Conventional PCR assays require standard gel electrophoresis for analysis, which increases the risk of false positive results due to amplicon carry-over. Real-time PCR is faster and there is no need for post-amplification analysis (Salisch et al., Citation1998). M. gallisepticum and M. synoviae PCR (MGMS PCR) assays have been applied for the single detection of M. gallisepticum (Carli & Eyigor, Citation2003; Mekkes & Feberwee, Citation2005; Callison et al., Citation2006; Grodio et al., Citation2008) and the simultaneous detection of both M. gallisepticum and M. synoviae (Jarquin et al., Citation2009). A duplex format has a huge advantage over monoplex formats, as it is particularly economical for small volume samples and for high-throughput laboratories in terms of cost of materials and turnaround time for both pathogens (Edwards & Gibbs, Citation1994). However, the duplex PCR assay for the simultaneous detection of M. gallisepticum and M. synoviae developed by Jarquin et al. (Citation2009) utilizes SYBR green dye that binds to all double-stranded DNAs, which is less specific. In addition, the primers in this assay for M. gallisepticum detection were previously shown to fail to distinguish between M. gallisepticum and Mycoplasma imitans (Garcia et al., Citation2005), so that it cannot be used in birds other than commercial chickens and turkeys (Dupiellet, Citation1984; Buntz et al., Citation1986; Bradbury et al., Citation1993). In case of need for extra specificity, TaqMan technology is preferred. Moreover, according to the Office International des Epizooties requirements, each PCR assay is recommended to incorporate an internal control (IC) (OIE, Citation2008). Only the assay described by Grodio et al. (Citation2008) had one.
In this study we report the development and validation of a highly specific and sensitive duplex MGMS PCR with IC using TaqMan-labelled probes (MGMS assay) that target the mgc2 gene for the detection of M. gallisepticum and the vlhA gene of M. synoviae. Validation of the assay was completed with 260 and 27 pooled samples (tracheal swabs) from commercial chickens and turkeys, respectively, with potential M. gallisepticum and M. synoviae involvement and 42 samples (non-pooled palatine cleft swabs) from backyard geese and ducks. The duplex MGMS PCR was compared with the isolation of M. gallisepticum and M. synoviae, conventional PCR assays (gapA PCR for M. gallisepticum and 16S PCR for M. synoviae) and PCR assays described in literature (Lauerman, Citation1998; Garcia et al., Citation2005).
Materials and methods
DNA extraction
A NucleoS+ kit (Biokom, Moscow) was used for extraction that included elutant, lysis buffer, saline buffer and buffer for elution. Briefly, 100 µl swab suspension was incubated with 30 µl elutant and 350 µl lysis buffer at room temperature for 5 min. After incubation 350 µl of 85% ethanol saline buffer was added to the lysate, which was then centrifuged and the supernatant discarded. Washing was repeated twice following the manufacturer's recommendations. Nucleic acid was eluted with 100 µl of 0.1 M Tris phosphate buffer or deionized water. Extraction was performed using swabs immediately after swabbing, and then swab samples were stored at −80°C until needed.
Duplex MGMS PCR
The duplex MGMS PCR was performed using the 7500 Real Time PCR System (Applied Biosystems, USA). Reagents for MGMS PCR with ROX reference dye (Syntol, Moscow) were used for the reaction. The final amount of the reaction mixture was 25 µl (according to the manufacturer's instructions), containing 12 pmol forward and reverse primers each for M. gallisepticum, 20 pmol forward and reverse primers each for M. synoviae, 5 pmol forward reverse primers each for IC, with corresponding probes (5 pmol each probe per reaction), IC template (added before extraction in an amount to allow for loss during extraction to correspond to 5.1×105 copies/reaction), 2.5 µl kit buffer per reaction (500 mM KCl, 150 mM HCl, 0.5% glycerin, 0.1% Tween and ROX), 0.5 µl Taq DNA polymerase (5 u/µl), 10 µl DNA sample and water to volume. After the initial denaturation at 95°C for 5 min, the duplex MGMS PCR was run with the following conditions for 40 cycles: at 95°C for 25 sec, at 58°C for 35 sec, and at 7°C for 45 sec. For all samples tested, any MGMS reaction that had a recorded the threshold cycle number (Ct) value was considered positive, and any MGMS reaction that had no recorded Ct value was considered negative. Statistical validation of Ct values was estimated with STATISTICA v. 6 software (StatSoft, Inc.).
Interpretation of real-time PCR
During amplification, the reporter dye (FAM, Cy5 and TAMRA) was measured against the passive reference dye (ROX) signal to normalize non-PCR-related fluorescence fluctuations occurring during the cycles. A positive result was assessed by identifying CT at which normalized reporter dye emission raised above background noise. If the fluorescent signal did not increase within 40 cycles, the sample was considered negative.
Primers and probes
The primers and probes utilized are presented in . For the detection of M. gallisepticum, primers and probe sequences were selected from the mgc2 fragment (AY556229) with Oligo 6.62 software (Woiciech and Piotr Rychlik, version 6.62; Molecular Biology Insights, Inc., USA). Primers mgc2-F and mgc2-R flank a 94 bp DNA segment. Fluorogenic mgc2 probe was labelled at the 5′ end with Cy5 reporter dye and approximately in the middle with black hole quencher (BHQ) as quencher.
Table 1. Primers and probes used in the assay
For the detection of M. synoviae, primers and probe sequences were selected from the vlhA fragment (AF035624) with Oligo 6.62 software. Primers vlhA-F and vlhA-R flank a 102 bp DNA segment. Fluorogenic vlhA probe was labelled at the 5′ end with FAM (6-carboxyfluorescein) reporter dye and approximately in the middle with BHQ as quencher.
For the detection of IC, primers and probe sequences were selected from the S3 gene fragment of avian reovirus (U 20642) with Oligo 6.62 software. Primers I-CF and IC-R flank a 104 bp DNA segment. Fluorogenic IC probe was labelled at the 5′ end with TAMRA (6-carboxytetramethylrhodamine) reporter dye and approximately in the middle with BHQ as quencher. Selected primers and probes were synthesized by Syntol Company (Moscow, Russia). Cross-reactivity of oligonucleotides was assessed by BLASTn analyses.
Sensitivity and reproducibility of the real-time MGMS PCR assay
The sensitivity of the duplex MGMS PCR was determined using seven 10-fold dilutions of extracted DNA of M. gallisepticum Rlow strain, starting with a titre of pure culture 7.0×106 colony-forming units (CFU)/ml and extracted DNA of M. synoviae WVU1853 strain, starting with a titre of pure culture 1.0×105 CFU/ml. The highest sample dilution yielding a Ct value was considered the detection limit of the duplex MGMS PCR, and the sensitivity was expressed in CFU equivalents per millilitre. Three independent runs of 10-fold dilutions of genomic DNA were carried out for the determination of mean Ct values and to evaluate reproducibility. Moreover, different concentrations of template DNA from each organism were mixed and tested by duplex MGMS PCR to ascertain the ability of this assay to co-amplify gene targets present in different relative amounts. Sensitivity was expressed in CFU equivalents per millilitre or DNA copies per reaction similar to the method described previously (Ge et al., Citation2001; Papazisi et al., Citation2003; Vasconcelos et al., Citation2005)
Specificity of the real-time MGMS PCR assay
Organisms used to assess the specificity of the assay are presented in . For each pathogen, the duplex MGMS PCR assay was performed individually. To determine the ability of the duplex MGMS PCR to reliably distinguish between M. gallisepticum and M. synoviae, it was tested with mixtures of DNAs of the microorganisms presented in in the following manner: mixture I, containing DNAs from four M. gallisepticum strains presented in ; mixture II, containing DNAs from M. synoviae strain WVU1853 and M. synoviae field isolate MS_V05 presented in ; mixture III, containing DNAs from M. gallisepticum strains, M. synoviae strain WVU1853 and M. synoviae field isolate presented in ; mixture IV, containing DNAs from all of the microorganisms presented in except for M. gallisepticum strains, M. synoviae strain WVU1853 and M. synoviae field isolate MS_V05; and mixture V, containing DNAs from all of the microorganisms presented in . The duplex MGMS PCR was also tested with serologically negative birds (Group A, ) and with palatine cleft swab pools (Group E, ).
Table 2. Bacterial and virus reference strains used for reaction specificity testing
Table 3. Samples tested for the duplex MGMS PCR validation
Internal control construction
A 600 bp product of the chicken reovirus S3 gene fragment was cloned into pUC18 plasmid vector (pGEM-T Easy; Promega, USA) according to the manufacturer's instructions. The plasmid DNA was extracted using GeneJet kit (Promega). Clones containing the proper insert were verified by sequencing. Purified plasmid DNA from one clone containing the proper insert was utilized. The concentration of the resulting plasmid was determined with a SmartSpec™ Plus spectrophotometer (BioRad, USA).
Laboratory and field samples
All field samples as tracheal swabs were collected from live layers and broilers exhibiting moderate to severe clinical signs of mycoplasma infection (airsacculitis, tracheal râles, nasal discharge, synovitis, poor performance) from various Russian poultry farms in 2005 to 2009. The birds' age and type of samples for each group are presented in . A total of 250 chickens for the experimental infection and 287 flocks with potential M. gallisepticum or M. synoviae infection (from five to 10 birds were sampled per flock) were examined for the validation of the duplex MGMS PCR.
Birds for the experiment originated from 7-day-old specific pathogen free embryos (White Leghorn). The birds were hatched and reared until 6 weeks of age in isolation units. The birds were housed at our Poultry Research Center at the Federal Center for Animal Health, (Vladimir, Russia). Feed and water were provided ad libitum throughout the experiment. The serologically negative status for M. gallisepticum and M. synoviae was determined prior to the experiment. Prior to infection and 14 days post inoculation, chickens were bled for serum. Serum samples were tested for M. gallisepticum and M. synoviae antibodies by the ProFlock MG and MS ELISA kits, respectively (Synbiotics, France). Prior to infection and at 14 days post inoculation, tracheal swab samples were collected from all of the birds with sterile, premoistened, cotton-tipped applicators (Copan, Italy). After being taken, each palatine swab was swirled in 1 ml sterile 0.1 M Tris-phosphate buffer, pH 7.4, squeezed completely, and then discarded. Negative control birds were given 0.5 ml sterile 0.1 M Tris-phosphate buffer, pH 7.4, as a negative control and kept in a separate room. No difference between sterile Frey broth and 0.1 M Tris-phosphate buffer was observed when inoculated into control birds (data not shown). All experiment groups (this one and those described below) were housed in Impedance isolation units (Impedance, Moscow).
Group A (non-inoculated) included 80 chickens serologically negative for M. gallisepticum and M. synoviae. A total of 80 birds were bled for serology. A total of 80 tracheal swabs were taken at 6 weeks.
Group B originated from 40 chickens serologically negative for M. gallisepticum and M. synoviae plus a negative control group (10 birds). The 40 chickens were allotted into groups of 20 each. The two groups were infected with 0.5 ml of 106 CFU inoculum (Frey broth) of M. gallisepticum (Rlow) intranasally and orally.
Group C originated from 40 White Leghorn birds, 4 weeks old, serologically negative for M. gallisepticum and M. synoviae plus a negative control group (10 birds). The 40 chickens were allotted into groups of 20 each. The two groups were infected with 0.5 ml of 105 CFU inoculum of M. synoviae (pathogenic M. synoviae field isolate MS_V05) intranasally and orally.
Group D originated from 40 White Leghorn birds, 4 weeks old, serologically negative for M. gallisepticum and M. synoviae plus a negative control group (10 birds). The 40 chickens were allotted into groups of 20 each. The two groups were infected both with 0.5 ml of 106 CFU inoculum of M. gallisepticum (Rlow) and 0.5 ml of 105 CFU inoculum of M. synoviae (pathogenic M. synoviae field isolate MS_V05) intranasally and orally.
Group E originated from 141 broiler flocks, 119 layer flocks and 27 turkey flocks with signs of mycoplasma infection. From five to 10 birds per flock were sampled. From five to 10 swab samples were pooled to comprise one pooled swab sample per flock. A total of 260 and 27 pooled samples (tracheal swabs) from commercial chickens and turkeys, respectively, with potential M. gallisepticum and M. synoviae involvement and 42 samples (non-pooled palatine cleft swabs) from backyard geese (35 non-pooled swabs) and ducks (seven non-pooled swabs) were used for PCR and isolation.
Analytical specificity and analytical sensitivity of the real-time duplex assay
The analytical specificity for M. gallisepticum (Asp MG) or for M. synoviae (Asp MS) of the duplex MGMS PCR for M. gallisepticum and M. synoviae, respectively, was determined using samples from M. gallisepticum-negative and M. synoviae-negative birds (Group A, ). The Asp was calculated with the formula Asp = true negative (TN) / (TN + false positive [FP]). The standard for calculating the Asp was conventional PCRs (gapA PCR for M. gallisepticum and 16S PCR for M. synoviae). Therefore, TNs were samples negative by gapA or 16S PCR and negative by the MGMS PCR. FPs were samples negative by gapA PCR or 16S PCR and positive by the duplex MGMS PCR.
The analytical sensitivity for M. gallisepticum (Asn MG) or for M. synoviae (Asn MS) of the duplex MGMS PCR was determined using samples from birds with potential M. gallisepticum or M. synoviae involvement (Group E, ). The Asn was calculated with the formula Asn = true positive (TP) / (TP + false negative [FN]). Isolation of M. gallisepticum and M. synoviae was used for calculation of Asn. Therefore, TPs were samples positive by isolation and by the duplex MGMS PCR assay. FNs were samples positive by isolation and negative by the MGMS PCR. The analytical sensitivity of the gapA PCR and 16S PCR as compared with M. gallisepticum and M. synoviae isolation, respectively, was also determined for the same group of samples (Group E, ).
Correlation of the duplex MGMS PCR assay with conventional gapA PCR for M. gallisepticum and 16S PCR for M. synoviae
The gapA PCR and 16S PCR assays were performed as previously described (Lauerman, Citation1998; Garcia et al., Citation2005) to assess the correlation between the MGMS PCR and the conventional gapA PCR and 16S PCR assays testing field samples. A total of 260 and 27 pooled samples (tracheal swabs) from commercial chickens and turkeys, respectively, with potential M. gallisepticum and M. synoviae involvement were tested by the three assays—that is, duplex MGMS PCR, gapA PCR and 16S PCR.
Testing of the MGMS PCR assay on samples from ducks and geese
To ensure the capability of the MGMS PCR to be used in avian species other than chickens and turkeys, a total of 42 palatine cleft swab samples (non-pooled) from backyard geese and ducks were screened for M. gallisepticum and M. synoviae at random (Vladimir region, Russia). Isolation of M. gallisepticum and M. synoviae was chosen as the gold standard. For comparison, gapA PCR (Garcia et al., Citation2005) for M. gallisepticum and 16S PCR (Lauermann, 1998) for M. synoviae were utilized for the same group of the samples.
PCR inhibition
Samples demonstrating PCR inhibition were treated as follows: storage at 4°C overnight and 1:10 dilution of the processed sample were used for removal of PCR inhibition.
Isolation of M. gallisepticum and M. synoviae
Isolation and identification of M. gallisepticum and M. synoviae were executed on tracheal swabs collected from flocks with signs of mycoplasma infection following standard protocols. Briefly, tracheal swabs were resuspended in 2 ml Frey's broth: 22.5 g/ml PPLO broth (BBL, USA), 50 ml yeast extract 25% (Sigma, Germany), 150 ml swine serum (Lesnoy, Russia), 5 ml thallium acetate 10% (Sigma, Germany), 12.5 ml NADH 1% (Serva, Germany), 3 g/ml glucose, 2.5 ml penicillin 500000 E and phenol red 1% and 12.5 ml cysteine (for M. synoviae) 1% (Labtech, Russia). Then 1 ml was incubated at 37°C until the broth medium colour indicator changed, and broth cultures were inoculated on Frey's media agar plates. Plates were incubated at 37°C and examined for the presence of mycoplasma colonies. M. gallisepticum and M. synoviae colonies were identified by the immunofluorescence technique described previously (Rosendal & Black, Citation1972).The other 1 ml was processed for PCR.
Colony counting
Twenty microlitre volumes of serial 10-fold dilutions were spread on mycoplasma agar plates in duplicate, incubated at 37°C under 5% carbon dioxide and examined weekly for the presence of colonies. Final colony counts were carried out after 14 days of incubation and the number of CFU per millilitre was calculated using only dilutions with less than 300 colonies per plate. All counts were expressed as log10 M. gallisepticum or M. synoviae CFU per millilitre.
Serology
Serum samples were collected from the negative, experimentally inoculated birds housed at our Research Center (Groups A, B, C and D; ) and naturally infected birds (Group E) submitted to the laboratory. Serum samples were tested for M. gallisepticum and M. synoviae antibodies by the ProFlock MG and MS ELISA kits, respectively (Synbiotics, France). Serologic tests for the identification of the respective agents were carried out in our laboratory. ELISA results were interpreted following the manufacturer's recommendations.
Results
Determination of specificity
The specificity of the duplex MGMS PCR was verified by testing DNA extracts from different avian pathogens, including M. gallisepticum and M. synoviae. All four M. gallisepticum strains, M. synoviae WVU1853 and M. synoviae field isolate tested were readily detected in the duplex MGMS PCR and in the conventional PCRs (). The duplex MGMS PCR was able to detect M. gallisepticum and M. synoviae independently and simultaneously (). The MGMS PCR tests detected none of the other bacteria tested. All of the M. gallisepticum strains tested produced recorded Ct values for Cy5 dye. The WVU1853 strain and the field isolate also produced recorded Ct values for FAM dye. The MGMS PCR exclusively amplified DNA simultaneously and/or individually from M. gallisepticum and M. synoviae-specific sequences in a single reaction (). When tested with serologically negative birds (Group A) the MGMS PCR produced only negative results, thus confirming its high specificity. Amplification reaction products were specific and corresponded to their lengths as predicted when visualized on an agarose gel (data not shown). No spurious PCR amplification reactions between the two mycoplasmas were noticed with various amounts of DNA.
Table 4. Testing of the analytical specificity of the duplex MGMS PCR in mixed samples
Sensitivity and reproducibility
The sensitivity for M. gallisepticum was determined using genomic template DNA extracted from seven 10-fold dilutions of M. gallisepticum Rlow strain, starting with a titre of 1×106 CFU/ml. The sensitivity for M. synoviae was evaluated using genomic DNA extracted from seven 10-fold dilutions of M. synoviae WVU1853 strain starting with a titre of 1×106 CFU/ml. The cultures of M. gallisepticum and M. synoviae were harvested at 48 h. Three replicates were carried out to determine the mean Ct values for each template. The average Ct values obtained for each reaction were plotted against the log10 of dilution and linear equations for M. gallisepticum and M. synoviae, respectively (y MG=0.291x – 4.89 and y MS=0.287x – 5.87) with RMG=0.999 and RMS=0.991 were generated. The duplex MGMS PCR maintained more obvious linearity for M. gallisepticum than for M. synoviae (). Using the slopes from the linear equations, the overall efficiencies of the duplex MGMS PCR for M. gallisepticum and M. synoviae were determined to be 95.43% and 93.82%, respectively ( and ; ). The detection limit of the duplex MGMS PCR was 7.0 and 1.0 CFU equivalents/ml for M. gallisepticum and M. synoviae, respectively, when tested independently or simultaneously, which corresponds to 29 genome copies and 34 genome copies per reaction for M. gallisepticum and M. synoviae, respectively (Ge et al., Citation2001; Papazisi et al., Citation2003; Vasconcelos et al., Citation2005). A mixture of M. gallisepticum and M. synoviae genomic DNA in various relative concentrations was amplified in a single reaction. Ct values (i.e. the detection limit) remained unchanged for the target that was in a lower relative concentration, even in the presence of a large amount (more than 1000-fold) of DNA from the microorganism that was co-amplified in the same tube (data not shown). Thus no reduction of sensitivity of the duplex MGMS PCR was observed.
Figure 1. Reproducibility and linearity of the duplex MGMS PCR for (1a) M. gallisepticum dilutions and (1b) M. synoviae dilutions. The assay standard curve was generated by plotting the Ct values versus log10 of 10-fold serial dilutions of extracted DNA of M. gallisepticum and M. synoviae, respectively. An efficiency of 95.43% for M. gallisepticum and 93.82% for M. synoviae, respectively, was estimated using the standard curve slope as indicated by the formula E=(10slope – 1)×100.
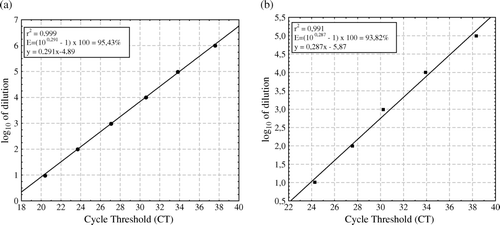
Table 5. Ct values obtained in three repeated duplex MGMS PCRs for M. gallisepticum
Table 6. Ct values obtained in three repeated duplex MGMS PCRs for M. synoviae
The reaction reproducibility was determined using the standard deviation of Ct values for each dilution. Standard deviations for M. gallisepticum and M. synoviae ranged from 0.036 to 0.53 and from 0.095 to 0.93, respectively. The presence of IC did not interfere with the amplification of mycoplasma targets throughout the cycles as compared with the absence of IC; that is, Ct values remained unchanged (data not shown).
Analytical specificity of the duplex MGMS PCR
Serum samples collected at preinoculation from Group A birds were all negative by the ProFlock MG and MS ELISA kits (Synbiotics, France). All tracheal swabs (Group A) collected at 6 weeks of age from the same group of birds were negative by the duplex MGMS PCR. All tracheal swabs (Group A) collected from the same group of birds were also negative by the gapA PCR and 16S PCR. Using the gapA PCR and 16S PCR as standards, the calculated Asp for the duplex MGMS PCR was 1.00. Additionally, serum samples were collected at 2 weeks after the experiment and all these were confirmed to be negative.
Detection of M. gallisepticum and/or M. synoviae in experimentally inoculated birds by the duplex MGMS PCR
In birds experimentally inoculated with M. gallisepticum and/or M. synoviae (Groups B, C and D), M. gallisepticum and M. synoviae could be detected 14 days after inoculation in tracheal swabs by the duplex MGMS PCR in a single tube (). The results of serologic tests at 14 days post inoculation serology gave positive results as follows: there were 25 positives for M. gallisepticum in Group B, 19 positives for M. synoviae in Group C, and 23 positives for M. gallisepticum and 18 positives for M. synoviae in Group D. The results of the duplex MGMS PCR at 14 days after inoculation demonstrated detection of M. gallisepticum and M. synoviae: in Group B, 100% of palatine swabs were positive only for M. gallisepticum by duplex MGMS PCR; in Group C, 100% of palatine swabs were positive only for M. synoviae by duplex MGMS PCR. In Group D, 100% of palatine swabs were positive simultaneously for M. gallisepticum and M. synoviae (). In relation to Groups B and C, neither PCR nor serology detected M. gallisepticum in any group other than Group B and neither PCR nor serology detected M. synoviae in any group other than Group C. All negative control birds in all of the groups tested negative for M. gallisepticum and M. synoviae both by the duplex MGMS PCR and serology.
Table 7. Detection of M. gallisepticum (MG) and M. synoviae (MS) and their antibodies prior to infection and 14 days post infection by the duplex MGMS PCR and ELISA
Analytical sensitivity and correlation of the duplex MGMS PCR assay with isolation and gapA PCR for M. gallisepticum and 16S PCR for M. synoviae
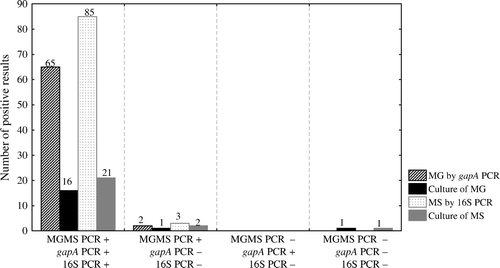
Results from birds with signs of mycoplasma infection (Group E, ) tested by the duplex MGMS PCR, gapA PCR and 16S PCR and isolation are shown in . From the 287 sample pools collected from birds with signs of infection, a total of 120 samples were positive by the duplex MGMS PCR assay, gapA PCR and 16S PCR assays, of which 35 samples were positive for M. gallisepticum by the duplex MGMS PCR and gapA PCR, and 55 samples were positive for M. synoviae by the duplex MGMS PCR and 16S PCR. The simultaneous result for M. gallisepticum and M. synoviae was obtained in 30 samples that were also confirmed by gapA PCR and 16S PCR. The isolation was successful in 16 and 21 samples for M. gallisepticum and M. synoviae, respectively. Twenty-four samples were negative both for M. gallisepticum and M. synoviae, and 59 samples were overgrown by secondary microorganisms. Two samples were duplex MGMS PCR assay-positive and gapA PCR-negative, but M. gallisepticum isolation was positive only in one sample. The isolation of M. gallisepticum in the second sample was unsuccessful because the culture was overgrown by secondary microorganisms. Three samples were duplex MGMS PCR assay-positive and 16S PCR assay-negative; M. synoviae isolation confirmed the presence of M. synoviae only in two samples, and in the third sample isolation of M. synoviae was unsuccessful because of overgrowth of secondary microorganisms. No sample was duplex MGMS PCR assay-negative and gapA PCR-positive or 16S PCR-positive. One hundred and sixty-two samples tested negative by the three PCRs, from which one sample tested positive by isolation for M. gallisepticum and one sample for M. synoviae, and 131 cultures were negative by isolation for M. gallisepticum and M. synoviae; isolation of M. gallisepticum and M. synoviae was unsuccessful in 29 samples because of overgrowth of secondary bacteria. Using M. gallisepticum and M. synoviae isolation for calculation, the Asn for the duplex MGMS PCR and gapA PCR was 0.944 and 0.941, respectively, and the Asn for the duplex MGMS PCR and 16S PCR was 0.958 and 0.954, respectively. When compared with each other, the duplex MGMS PCR and gapA PCR agreed in 94.5%. When compared with each other, the duplex MGMS PCR and 16S PCR correlated in 96.6%.
Testing of the duplex MGMS PCR assay on samples from backyard ducks and geese
A total of 42 non-pooled palatine cleft swab samples from backyard ducks and geese were tested by the duplex MGMS PCR, gapA PCR, 16S PCR, and isolation. All samples tested negative by the duplex MGMS PCR assay with 100% agreement with gapA PCR and 16S PCR assays except for five samples, three of which were M. gallisepticum-positive and two were M. synoviae-positive. Five positives for M. gallisepticum and two positives for M. synoviae were also confirmed by gapA PCR and 16S PCR, respectively. Isolation of M. gallisepticum and M. synoviae was negative in all samples (i.e. no culture was grown).
Application of internal control
The IC reaction (reovirus origin) was used as a control to screen for inhibition of amplification. The IC reaction is supposed to be positive whatever samples for M. gallisepticum and M. synoviae are tested. All samples from serologically negative birds (Group A, ) while testing the analytical sensitivity, samples containing genomic DNA of the microorganisms presented in while testing the specificity, samples for testing the detection limit ( and ), and all samples from experimentally infected birds (Groups B, C and D) were IC-positive (data not shown). In testing samples from naturally infected birds (Group E, ), the IC reaction was negative in 12 samples out of 287. These 12 samples were also negative for M. gallisepticum and M. synoviae by the duplex MGMS PCR, gapA PCR and 16S PCR. Upon re-testing, the IC reaction in those samples was positive in all 10 samples, from which one sample was M. gallisepticum-positive and M. synoviae-positive and one sample was M. gallisepticum-positive. After re-testing of those 12 samples, the agreement between the duplex MGMS PCR and gapA PCR and 16S PCR was 100%. Isolation of M. gallisepticum and M. synoviae was unsuccessful in all 12 samples because of overgrowth of secondary bacteria.
Testing the palatine cleft swab samples from ducks and geese, the IC reaction was positive in 37 out of 42 samples. Five IC reactions were inhibited. Storage at 4°C overnight and 1:10 dilution of these processed samples removed inhibition; however, the five samples proved to be negative for M. gallisepticum or M. synoviae.
Discussion
The primary goal of the current study was to develop a real-time duplex TaqMan PCR assay with an incorporated IC. The assay can be applied to the simultaneous detection M. gallisepticum and M. synoviae. Using this duplex MGMS PCR assay, detection of both mycoplasmas was possible in a single-tube reaction, which considerably speeds up the identification of these pathogenic agents. Our assay was able to simultaneously reveal M. gallisepticum and M. synoviae-specific sequences from different strains in the background of DNA extracts from related and non-related bacteria capable of colonizing the avian respiratory tract. The designed primers were checked by BLASTn analyses for cross-reactivity and potential of non-specific annealing. Results of the BLASTn analyses showed that the M. synoviae probe and M. synoviae reverse primer shared significant homology with the pMGA1.1 precursor for M. gallisepticum. However, this did not compromise PCR and amplification in all cases is specific because the M. synoviae forward primer is highly specific only to M. synoviae sequences. The M. synoviae probe undergoes cleavage only when Taq DNA polymerase amplifies the M. synoviae forward primer. Therefore, non-specific probe interaction is not detectable with non-amplifiable product. The other primer and probe sequences were highly specific.
The duplex MGMS PCR could be used for screening not only commercial chickens and turkey flocks but also ducks and geese, whereas assays based on the 16S rRNA gene for M. gallisepticum detection cannot be applied (Mekkes & Feberwee, Citation2005; Jarquin et al., Citation2009). When tested with serologically negative birds (Group A) the duplex MGMS PCR produced only negative results for M. gallisepticum and M. synoviae, thus confirming its high specificity. The assay's detection limit with dilution of template DNA was 7 and 1 CFU equivalents/ml for M. gallisepticum and M. synoviae, respectively, which corresponds to 29 and 34 DNA copies per reaction.
Considering conventional PCRs and isolation as the standards for the diagnosis of M. gallisepticum and M. synoviae infections, the calculated Asp MG and Asp MS for the duplex MGMS PCR assay were 1.00 and 1.00, respectively, whereas Asn MG and Asn MS were 0.94 and 0.95, respectively. One hundred and forty-one broiler flocks, 119 layer flocks and 27 turkey flocks were sampled. A total of 141 plus 119 and 27 pooled samples from commercial chickens and turkeys, respectively, with potential M. gallisepticum and M. synoviae involvement were analysed by the duplex MGMS PCR, producing 37 positive results for M. gallisepticum, 58 positive results for M. synoviae and 30 samples that tested positive for M. gallisepticum and M. synoviae simultaneously. In analysing the field samples in comparison with the conventional reference assays in terms of correlation, the overall agreement between the duplex MGMS PCR and gapA PCR plus duplex MGMS PCR and 16S PCR was 94.5% (65 samples) and 96.6% (85 samples), respectively. These results obtained by the duplex MGMS PCR and the reference PCRs are highly correlated. However, multiplex real-time PCR methods offer an attractive alternative to conventional PCR in the diagnostic laboratory. Results are available within several hours in a highly standardized format without handling of PCR products, which subsequently decreases the risk of false positive results due to amplicon carry-over. Furthermore, this technique offers the possibility to detect multiple pathogens simultaneously, which is particularly beneficial for high-throughput laboratories. For instance, in the absence of a duplex PCR assay, samples for M. gallisepticum and M. synoviae must be tested by two PCR assays, which is time-consuming and labour intensive. In addition, incorporation of an IC provides an advantage for monitoring inhibitory substances present in extracted DNA or reagent breakdown. Samples to test could be spiked with IC with a known concentration before extraction. The Ct value of IC in PCR will be very informative in terms of a proper extraction procedure and inhibitory effects that can lead to the failure of PCR to detect a low copy number of the target. Using quantified IC copies per reaction, discrepancy between the expected Ct values of IC and the actual ones could be easily observed (data not shown). It is noteworthy that false-negative results are highly likely to occur (Carli & Eyigor, Citation2003). Due to inclusion of IC in the field samples, we did identify 12 samples whereby amplification reaction was inhibited. Re-testing revealed two positive results for M. gallisepticum and one positive result for M. synoviae. If IC, had not been used, these positives would have been missed. As IC we used plasmid DNA, which was chosen over genomic DNA in light of the fact that it is more stable and can be accurately quantified.
At present, the use of conventional PCR and real-time PCR to detect M. gallisepticum, M. synoviae and M. gallisepticum plus M. synoviae has been utilized (). None of the assays have been validated in birds other than commercial chickens. Some authors attempted to assess sensitivity. For this purpose, they used pure culture (Carli & Eyigor, Citation2003; Mekkes & Feberwee, Citation2005: Jarquin et al., Citation2009), plasmid (Callison et al., Citation2006) and amplicon copies of targets (Raviv & Kleven, Citation2009), and obtained varied values for their assays. The lower sensitivity of our assay for the M. gallisepticum detection in comparison with that of the assays previously described—0.01 to 0.06 CFU equivalents/ml (Mekkes & Feberwee, Citation2005) and 3.3×103 CFU/ml (Carli & Eyigor, Citation2003)—is probably attributable to the use of TaqMan probes in the current assay. TaqMan-based real-time assays for the single detection of M. gallisepticum were designed with the detection limit of 25 and 14 plasmid copies containing a specific gene insert of M. gallisepticum per reaction (Callison et al., Citation2006; Grodio et al., Citation2008). The sensitivity for M. gallisepticum in duplex PCR by Jarquin et al. (Citation2009) was similar, being down to 25 to 27 gene copies per reaction and for M. synoviae being 28 gene copies. Raviv & Kleven (Citation2009) reported two real-time Taq-Man PCR assays for M. gallisepticum and M. synoviae detection with an extreme sensitivity of 1 and 10 DNA copies per reaction using as a standard quantified amplicon of the genomic regions targeted by the two assays. However, none of the reported assays except for that of Grodio et al. (Citation2008) included an IC in the PCR. They utilized an IC for correlating conjunctivitis with M. gallisepticum load. However, only one assay by Jarquin et al. (Citation2009) was in a duplex format based on the 16S rRNA genes for detection of M. gallisepticum and M. synoviae utilizing SYBR green technology that is less specific than TaqMan. Specificity and sensitivity are a great concern when M. gallisepticum or M. synoviae are suspected in flocks. TaqMan probe technology gives an extra level of specificity unlike SYBR Green dye. Thus, the sensitivity of our assay was 7 and 1 CFU/ml for M. gallisepticum and M. synoviae, respectively, which corresponds to 29 and 34 DNA copies per reaction (Ge et al., Citation2001)–which is less than that of Raviv & Kleven (Citation2009). Although direct comparison of detection limits between assays cannot be made. Hess et al. (Citation2007) reported on comparison of PCR assays to detect M. gallisepticum and M. synoviae on dry swab samples. The detection limit of both real-time and conventional assays being used for M. gallisepticum and M. synoviae detection was down to 100 DNA copies per reaction. However, the assays reported by Hess et al. (Citation2007), Mekkes & Feberwee (Citation2005) and Jarquin et al. (Citation2009) cannot distinguish between M. gallisepticum and M. imitans (Garcia et al., Citation2005), and therefore cannot be used in ducks and geese. This is a huge drawback as M. gallisepticum and M. synoviae has been reported to be found in ducks, geese and peafowl (Buntz et al., Citation1986; Benčina et al., Citation1988; Cookson & Shivaprasad, Citation1994). Among the reported real-time assays for M. gallisepticum detection, only the assay by Raviv & Kleven (Citation2009) is free of cross-reaction with M. imitans. Following on from this, a highly specific PCR assay capable of detecting M. gallisepticum and M. synoviae in a wider variety of avian species would be very helpful. The assay we report on can potentially serve as an additional tool for screening not only chicken and turkey flocks, but also duck and geese flocks for M. gallisepticum and M. synoviae in a field environment. The duplex MGMS PCR is slightly less sensitive than that of Raviv & Kleven (Citation2009) but it is as highly specific. Moreover, it combines the capability of detecting two targets at once. The validation of our assay was completed by testing tracheal swab samples collected at random from backyard geese and ducks, which have not been included in such PCR assays hitherto. Forty-two samples were tested of which three and two samples were positive by the duplex MGMS PCR for M. gallisepticum and M. synoviae, respectively, whereas gapA PCR and 16S PCR assays also confirmed M. gallisepticum and M. synoviae in those samples with 100% agreement. Isolation of M. gallisepticum and M. synoviae was negative in all samples due to overgrowth of saprophytic bacteria. Such a case of prolific overgrowth has been reported previously in geese (Buntz et al., Citation1986) Overall, this is the first report on M. gallisepticum or M. synoviae in backyard poultry in Russia. All sampled ducks and geese were clinically healthy without any slight sign of infection.
Table 8. Comparison of various PCR assays for detection of M. gallisepticum and M. synoviae
In summary, we have developed a rapid, robust, sensitive and specific duplex MGMS PCR assay for the simultaneous detection of the important avian pathogens such as M. gallisepticum and M. synoviae in clinical samples from commercial and backyard poultry including ducks and geese. After reception of the sample, analysis could be completed within hours with high sensitivity. The duplex MGMS PCR was able to simultaneously amplify multiple gene targets in the same tube, being cost-effective and saving time in the diagnostic laboratory. The incorporation of an IC helps avoid false negative results. We hope that in the near future the duplex MGMS PCR will be used as a supplementary diagnostic tool for the detection of M. gallisepticum and M. synoviae in clinical samples from chickens, turkeys, ducks and geese, and probably other avian species.
Acknowledgements
This research was supported by the ISTC grant #3017 of ARS USDA. The authors thank Dr Jeff Evans for reading the manuscript. They are also grateful to the backyard farmers of Vladimir region for providing flocks for sampling.
References
- Avakian , A.P. and Kleven , S.H. 1990 . The humoral immune response of chickens to Mycoplasma gallisepticum and potential causes of false positive reactions in avian mycoplasma serology . Zentralblatt fur Bakteriologie Suppl , 20 : 500 – 512 .
- Benčina , D. , Tadina , T. and Dorrer , D. 1988 . Natural infection of ducks with Mycoplasma synoviae and Mycoplasma gallisepticum and mycoplasma egg transmission . Avian Pathology , 17 : 441 – 449 .
- Bradbury , J.M. 2005 . Poultry mycoplasmas: sophisticated pathogens in simple guise . British Poultry Science , 46 : 125 – 136 .
- Bradbury , J.M. , Abdul-Wahab , O.S.M. , Yavari , C.A. , Dupiellet , J.P. and Bove , J.M. 1993 . Mycoplasma imitans sp. nov. is related to Mycoplasma gallisepticum and found in birds . International Journal of Systematic Bacteriology , 43 : 721 – 728 .
- Buntz , B. , Bradbury , J.M. , Vuillaume , A. and Rousselot-Paillet , D. 1986 . Isolation of Mycoplasma gallisepticum from geese . Avian Pathology , 15 : 615 – 617 .
- Callison , S.A. , Riblet , S.M. , Sun , S. , Ikuta , N. , Hilt , D. Leiting , V. 2006 . Development and validation of a real-time TaqMan polymerase chain reaction assay for the detection of Mycoplasma gallisepticum in naturally infected birds . Avian Diseases , 50 : 537 – 544 .
- Carli , K.T. and Eyigor , A. 2003 . Real-time polymerase chain reaction for detection of Mycoplasma gallisepticum in chicken trachea . Avian Diseases , 47 : 712 – 717 .
- Cookson , K.C. and Shivaprasad , H.L. 1994 . Mycoplasma gallisepticum infection in chukar partridges, pheasants, and peafowl . Avian Diseases , 38 : 914 – 921 .
- Dupiellet J.P. 1984 Mycoplasmes et acholeplasmes des palmipedes a foie gras: isolement caractérisation, étude du rôle dans la pathologie Rappport de D.E.A. de Pathologie Université de Bordeaux II, Villenave d' Ornon, France
- Edwards , M.C. and Gibbs , R.A. 1994 . Multiplex PCR: advantages, development, and applications . Genome Research , 3 : 65 – S75 .
- Garcia , M. , Ikuta , N. , Levisohn , S. and Kleven , S.H. 2005 . Evaluation and comparison of various PCR methods for detection of Mycoplasma gallisepticum infection in chickens . Avian Diseases , 49 : 125 – 132 .
- Garcia , M. , Jackwood , M.W. , Levisohn , S. and Kleven , S.H. 1995 . Detection of Mycoplasma gallisepticum, Mycoplasma synoviae, and Mycoplasma iowae by multi-species polymerase chain reaction and restriction fragment length polymorphism . Avian Diseases , 39 : 606 – 616 .
- Ge , Z. , White , D. , Whary , M. and Fox , J. 2001 . Fluorogenic PCR-based quantitative detection of a murine pathogen, Helicobacter hepaticus . Journal of Clinical Microbiology , 35 : 2598 – 2602 .
- Georgiades , G. , Iordanidis , P. and Koumbati , M. 2001 . Cases of swollen head syndrome in broiler chickens in Greece . Avian Diseases , 45 : 745 – 750 .
- Grodio , J.L. , Dhondt , K.V. , O'Connell , P.H. and Schat , K.A. 2008 . Detection and quantification of Mycoplasma gallisepticum genome load in conjunctival samples of experimentally infected house finches (Carpodacus mexicanus) using real-time polymerase chain reaction . Avian Pathology , 37 : 385 – 391 .
- Hammond , P.P. , Ramírez , A.S. , Morrow , C.J. and Bradbury , J.M. 2009 . Development and evaluation of an improved diagnostic PCR for Mycoplasma synoviae using primers located in the haemagglutinin encoding gene vlhA and its value for strain typing . Veterinary Microbiology , 136 ( 1–2 ) : 61 – 68 .
- Hess , M. , Neubauer , C. and Hackl , R. 2007 . Interlaboratory comparison of ability to detect nucleic acid of Mycoplasma gallisepticum and Mycoplasma synoviae by polymerase chain reaction . Avian Pathology , 36 : 127 – 133 .
- Hong , Y. , García , M. , Leiting , L. , Benčina , D. , Dufour-Zavala , L. , Zavala , G. and Kleven , S.H. 2004 . Specific detection and typing of Mycoplasma synoviae strains in poultry with PCR and DNA sequence analysis targeting the hemagglutinin encoding gene vlhA . Avian Diseases , 48 : 606 – 616 .
- Jarquin , R. , Schultz , J. , Hanning , I. and Ricke , S. 2009 . Development of a real-time polymerase chain reaction assay for the simultaneous detection of Mycoplasma gallisepticum and Mycoplasma synoviae under industry conditions . Avian Diseases , 53 : 73 – 77 .
- Kempf , I. , Gesbert , F. and Guittet , M. 1997 . Experimental infection of chickens with an atypical Mycoplasma gallisepticum strain: comparison of diagnostic methods . Research in Veterinary Science , 63 : 211 – 213 .
- Khan , M.I. and Kleven , S.H. 1993 . Detection of Mycoplasma gallisepticum infection in field samples using a species-specific DNA probe . Avian Diseases , 37 : 880 – 883 .
- Kleven , S.H. 1975 . Antibody response to avian mycoplasmas . American Journal of Veterinary Research , 36 : 563 – 565 .
- Kleven , S.H. 1997 . “ Mycoplasma synoviae infection ” . In Diseases of Poultry , 10th edn , Edited by: Calnek , B.W. , Barnes , H.J. , Beard , C.W. , McDougald , L.R. and Saif , Y.M. 220 – 227 . Ames : Iowa State Press .
- Kleven , S.H. 1998 . “ Mycoplasmosis ” . In A Laboratory Manual for the Isolation and Identification of Avian Pathogen , 4th edn , Edited by: Swayne , D.E. , John , R. , Jackwood , M.W. , Pearson , J.E. and Reed , W.M. 100 – 105 . Kennett Square , PA : University of Pennsylvania, New Bolton Center .
- Kleven , S.H. , Jordan , F.T.W. and Bradbury , J.M. 1996 . “ Avian mycoplasmosis (Mycoplasma gallisepticum) ” . In Manual of Standards for Diagnostic Tests and Vaccines , Edited by: Reichard , R. 512 – 521 . Paris : Office International des Epizooties .
- Lauerman , L.H. 1998 . “ Mycoplasma PCR assays ” . In Nucleic Acid Amplification Assays for Diagnosis of Animal Diseases , Edited by: Lauerman , L.H. 41 – 52 . Davis , CA : American Association of Veterinary Laboratory Diagnosticians .
- Levisohn , S. and Kleven , S.H. 2000 . “ Avian mycoplasmosis (Mycoplasma gallisepticum) ” . In Diseases of Poultry: World Trade and Public Health Implications , Edited by: Beard , C.W. and McNulty , S. Vol. 19 , 425 – 442 . Paris : Office International des Epizooties .
- Ley D.H. 2003 Mycoplasma gallisepticum infection In Y.M. Saif H.J. Barnes J.R. Glisson A.M. Fadly L.R. McDougald D.E. Swayne Diseases of Poultry , 11th edn 722 744 Ames , IA Iowa State Press
- Ley , D.H. and Yoder , H.W. Jr . 1997 . “ Mycoplasma gallisepticum infection ” . In Diseases of Poultry , 10th edn , Edited by: Calnek , B.W. , Barnes , H.J. , Beard , C.W. , McDougald , L.R. and Saif , Y.M. 194 – 207 . Ames : Iowa State Press .
- Mardassi , B.B.A. , Mohamed , R.B. , Gueriri , I. , Boughattas , S. and Milk , B. 2005 . Duplex PCR to differentiate Mycoplasma synoviae and Mycoplasma gallisepticum on the basis of conserved species-specific sequences of their hemagglutinin genes . Journal of Clinical Microbiology , 43 : 948 – 958 .
- Marois , C. , Dufour-Gesbert , F. and Kempf , I. 2002 . Polymerase chain reaction for detection of Mycoplasma gallisepticum in environmental samples . Avian Pathology , 31 : 163 – 168 .
- Mekkes , D.R. and Feberwee , A. 2005 . Real-time polymerase chain reaction for the qualitative and quantitative detection of Mycoplasma gallisepticum . Avian Pathology , 34 : 348 – 354 .
- Nascimento , E.R. , Yamamoto , R. , Herrick , K.R. and Tait , R.T. 1991 . Polymerase chain reaction for detection of Mycoplasma gallisepticum . Avian Diseases , 35 : 62 – 69 .
- OIE 2008 OIE Quality Standard and Guidelines for Veterinary Laboratories: Infectious Diseases Paris Office international des Epizooties
- Pang , Y. , Wang , H. , Girshick , T. , Xie , Z. and Khan , M.J. 2002 . Development and application of a multiplex polymerase chain reaction for avian respiratory agents . Avian Diseases , 46 : 691 – 699 .
- Papazisi , L. , Gorton , T.S. , Kutish , G. , Markham , P.F. , Browning , G.F. Nguyen , D.K. 2003 . The complete genome sequence of the avian pathogen Mycoplasma gallisepticum strain Rlow . Microbiology , 149 : 2307 – 2316 .
- Ramírez , A.S. , Naylor , C.J. , Hammond , P.P. and Bradbury , J.M. 2006 . Development and evaluation of a diagnostic PCR for Mycoplasma synoviae using primers located in the intergenic spacer region and the 23S rRNA gene . Veterinary Microbiology , 118 : 76 – 82 .
- Raviv , Z. , Ferguson-Noel , N. , Laibinis , V. , Wooten , R. and Kleven , S.H. 2007 . Role of Mycoplasma synoviae in commercial layer Escherichia coli peritonitis syndrome . Avian Diseases , 51 : 685 – 690 .
- Raviv , Z. and Kleven , S.H. 2009 . The development of diagnostic real-time TaqMan PCRs for the four pathogenic avian mycoplasmas . Avian Diseases , 53 : 103 – 107 .
- Rosendal , S. and Black , F.T. 1972 . Direct and indirect immunofluorescence of unfixed and fixed mycoplasma colonies . Acta Pathologica et Microbiologica Scandinavica, B , 80 : 615 – 622 .
- Salisch , H. , Hinz , K.H. , Graack , H.D. and Ryll , M. 1998 . A comparison of a commercial PCR-based test to culture methods for detection of Mycoplasma gallisepticum and Mycoplasma synoviae in concurrently infected chickens . Avian Pathology , 27 : 142 – 147 .
- Vasconcelos , A.T. , Ferreira , H.B. , Bizarro , C.V. , Bonatto , S.L. , Carvalho , M.O. Pinto , P.M. 2005 . Swine and poultry pathogens: The complete genome sequences of two strains of Mycoplasma hyopneumoniae and a strain of Mycoplasma synoviae . Journal of Bacteriology , 187 : 5568 – 5577 .
- Wang , H. , Fadl , A.A. and Khan , M.I. 1997 . Multiplex PCR for avian pathogenic mycoplasma . Molecular and Cellular Probes , 11 : 211 – 216 .