Abstract
A rapid fowl adenovirus (FAdV) classification method based on a 30-bp sequence of the hexon loop (L1) was developed using the pyrosequencing technique. FAdV identification is relevant for epidemiological studies and for the adoption of a correct strategy where vaccination is to be used for the control of the disease. FAdV typing is usually performed using polymerase chain reaction coupled with either conventional DNA sequencing or restriction enzyme analysis; however, both methods can be time consuming and/or very expensive to be used as a routine tool. In the present study, polymerase chain reaction and subsequent pyrosequence analysis of the variable hexon L1 region were assessed in order to rapidly differentiate FAdV species. Forty-nine FAdV samples (22 reference strains and 27 field isolates) were tested and the results were compared with those obtained by conventional DNA sequencing. The results clearly demonstrated that pyrosequence analysis provides a new approach for a rapid differentiation and classification of the FAdV species that is faster, more cost-effective and easier to interpret than other techniques commonly used.
Introduction
Fowl adenoviruses (FAdVs), belonging to the Aviadenovirus genus of the family Adenoviridae, are distributed worldwide and known to cause significant economic problems in the poultry industry, mainly due to inclusion body hepatitis (IBH) and hydropericardium syndrome (McFerran & Connor, Citation1977; Christensen & Saifuddin, Citation1989; Nakamura et al., Citation1999; Toro et al., Citation1999; Hess, Citation2000; Alvarado et al., Citation2007), but are also associated with a wide range of clinical presentations including gizzard erosion (Ono et al., Citation2001, Citation2003; Muroga et al., Citation2006) and pancreatitis (Grimes et al., Citation1977; Nakamura et al., Citation2002).
FAdVs are mostly transmitted horizontally, being present in all excretions and at high titres in faeces. Vertical transmission is also possible where the virus can be passed through embryonated eggs and can be reactivated in chicks that are a few weeks old, especially if the birds are immunodepressed (McFerran & Smyth, Citation2000; Grgic et al., Citation2006).
Various serological tests, such as enzyme-linked immunosorbent assays (Hess, Citation2000; Balamurugan & Kataria, Citation2004; Philippe et al., Citation2007), agar gel immunodiffusion and immunofluorescence (Balamurugan & Kataria, Citation2004) can be used for antibody detection. However, since serological methods detect only common group-specific antigens, and because of the widespread occurrence of antibodies to avian adenovirus, these techniques are only of minor relevance for their diagnosis. For these reasons, investigations are mainly focused on direct detection and identification of the agent (Hess, Citation2000) that can be isolated both from sick and healthy birds (McFerran & Smyth, Citation2000).
FAdVs have been grouped into five species (A to E), based on their molecular structure, and divided into 12 serotypes, based on cross-neutralization assays (Hess, Citation2000).
Despite the fact that FAdVs are generally considered ubiquitous in poultry farms, certain species and serotypes are known to be associated with primary disease such as IBH, while others cause no clinical disease. Characterization of FAdVs isolated in Canada (Ojkic et al., Citation2008), New Zealand (Saifuddin & Wilks, Citation1991) and Australia (Erny et al., Citation1991) showed that the IBH syndrome is mainly associated with FAdV species E and D, while species A and C have been less frequently found in severe IBH cases (Toro et al., Citation1999; Ojkic et al., Citation2008). FAdV classification is therefore very useful for epidemiological tracing and is of critical importance where vaccination is to be used for the control of the disease (Toro et al., Citation2002; Jadhao et al., Citation2003; Balamurugan & Kataria, Citation2004; Steer et al., Citation2009).
The hexon protein is the major capsid protein of the non-enveloped virion on which type, group, and subgroup-specific determinants are located (Norby, Citation1969). The hexon gene consists of conserved regions (pedestals), which are located more inside the virion, and variable loops, which protrude from the surface (Roberts et al., Citation1986; Athappilly et al., Citation1994), containing the type-specific neutralizing epitopes (Toogood et al., Citation1992; Adam et al., Citation1998).Due to the interaction with the immune system, the sequence identity between loop regions of different species is low (Sheppard et al., Citation1995; Crawford-Miksza & Schnurr, Citation1996). The hexon loop 1 (L1) represents the most variable region and has proved to be the most useful region in identifying and differentiating FAdV species and serotypes, when used in polymerase chain reaction (PCR) coupled either with conventional DNA sequencing or with restriction fragment length polymorphism (RFLP) analysis (Raue & Hess, Citation1998; Hess et al., Citation1999; Meulemans et al., Citation2001; Steer et al., Citation2009). However, these methods are time consuming, often require extensive interpretation, and the running cost per analysis can be relatively high to be used as a routine typing tool.
Pyrosequencing has proven to be a rapid, sensitive and high-throughput method for sample screening and genotyping in different fields of disease research (Ronaghi & Elahi, Citation2002; Elahi et al., Citation2003; Swan et al., Citation2006; Deyde & Gubareva, Citation2009; Deyde et al., Citation2009; Quince et al., Citation2009). On the strength of these considerations, the aim of the present study was to develop a rapid FAdV species classification method, based on the pyrosequencing of a 30-bp L1 region of the hexon gene.
Materials and Methods
Viruses
Twenty-two FAdV reference strains (), representatives of currently recognized distinct serotypes, and 27 field isolates collected in Italy between 2007 and 2008 were used in this study.
Table 1. FAdV reference strains used in the present study.
All the viruses were grown in chicken embryo liver cells obtained from 14-day-old specific pathogen free embryos (Lohmann). Chicken embryo liver cells (1 x 106 cells/ml) in 25-cm2 culture flasks were incubated at 37°C with 5% carbon dioxide in Minimal Essential Medium Eagle (Sigma) containing 5% foetal calf serum (Invitrogen), L-glutamine 2 mM (Sigma) and 1% PenStrep (Sigma). Cell monolayers were inoculated with 500 µl viral suspension and, after 1 h absorption, were incubated for 2 to 3 days, until an extensive cytopathic effect could be seen. The infected cells and their supernatants were then harvested and freeze–thawed three times. The suspensions were centrifuged at 3000 x g for 10 min to eliminate cell debris and the supernatants were stored at –20°C.
Hexon L1 conventional PCR amplification
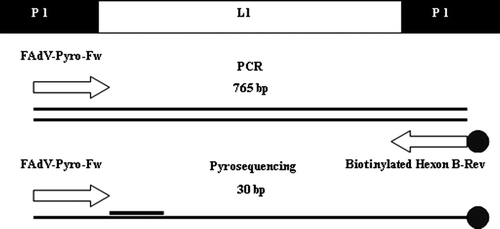
Hexon L1 Genbank sequences CELO AF339914, 685 AF508947, P7-A AF339915, SR49 AF508948, 75 AF508949, 75-1A-1 AF339921, J2-A AF339917, 506 AF508950, KR5 AF508951, TR22 AF508953, IBH-2A AF339916, CR119 AF508954, YR36 AF508955, X11-A AF339924, TR59 AF508956, 58 AF508957, T8-A AF339919, 764 AF508958, B-3A AF339922, A-2A AF339918, C-2B AF339923 and 380 AF339925 were aligned using MEGA software (version 4.1) in order to identify a 30-bp region capable of discriminating different FAdV species. The conserved sequence upstream of this 30-bp region was used to design the PCR forward primer FAdV-Pyro-fw (5'-TGGGTBYTGGACATGGG-3'). Adenoviral dsDNA was extracted using the Roche High Pure PCR Template Preparation Kit, according to the manufacturer's instructions. PCR was performed using the FadV-Pyro-fw primer and the reverse primer Hexon B (5'-TAGTGATGMCGSGACATCAT-3') (Meulemans et al., Citation2001), in order to generate a PCR product between 740 bp (FAdV-4: J2-A AF339917, 506 AF508950, KR5 AF508951; FAdV-10: C-2B AF339923) and 765 bp (FAdV-5: IBH-2A AF339916). DNA amplifications were carried out in a total volume of 50 µl containing 5 µl viral DNA, 5 pmol each primer, 1.5 mmol MgCl2, 0.8 mmol dNTPs and 5 units AmpliTaq Gold® (Roche). The PCR was started with an initial denaturation step of 10 min at 95°C. The temperature profile of the following 40 cycles consisted of 1 min at 95°C for denaturation, 1 min at 55°C for primer annealing and 90 sec at 72°C for elongation. The reaction was terminated by a final elongation step of 7 min at 72°C. The amplification products were separated on a 1.2% agarose gel stained with ethidium bromide and visualized by ultraviolet transillumination. The size of the amplification product was confirmed by comparison with a commercial PCR marker (Roche DNA Molecular Weight Marker VI 0.15—2.1 kbp) in agarose gel and by the following sequence analysis. PCR specificity was appraised against the DNA of haemorrhagic enteritis virus (HEV), egg drop syndrome virus (EDS'76), infectious laryngotracheitis virus (ILTV) and against the cDNA of infectious bronchitis virus (IBV serotype M41) and avian influenza virus (AIV serotype H7).
DNA sequencing and BLAST alignment
Hexon L1 PCR products were purified using High Pure PCR Product Purification (Roche) and sequenced using the same primers used for PCR and the Big Dye® Terminator v 3.1 Cycle sequencing Kit, according to the manufacturer's protocol (Applied Biosystems Inc.).
The sequences were analysed using an ABI PRISM 3130xl Genetic Analyzer (Applied Biosystems Inc.) and aligned using the BLASTn program (http://blast.ncbi.nlm.nih.gov/Blast.cgi) to confirm the identity of the viral strains. Aiming to verify the relationship between FAdV samples, phylogenetic analysis of the aligned sequences was performed and neighbour-joining distance trees together with the pairwise distance analysis were performed using the MEGA 4.1 program (Tamura et al., Citation2007).
Pyrosequencing PCR
PCR was performed using the same set of primers as for the previous PCR; however, the Hexon B primer was biotinylated (5'-[Biotag]TAGTGATGMCGSGACATCAT-3') (). DNA amplifications were carried out in a total volume of 50 µl using the same template DNA, reagents and cycling conditions as before, with the exception of the primer concentrations, which were increased to 15 pmol.
Pyrosequencing reaction
Amplification products were immobilized onto streptavidin-coated sepharose beads (5 µl/sample) and washed in a series of buffers according to Biotage's recommendations (PyroMark™ Sample Prep Guidelines; Biotage). Single-stranded biotinylated DNA products were hybridized to FAdV-Pyro-Fw sequencing primer in a 96-well plate used at a final concentration of 0.5 µM in 40 µl annealing buffer. Pyrosequencing was performed in a Pyromark ID platform pyrosequencer using Pyro Gold reagents according to the manufacturer's instructions. The related sequences were analysed and compared with a FAdV hexon sequences library using IdentiFire software (Biotage). The library was compiled using the same Genbank sequences used to design the FAdV-Pyro-Fw primer, with the addition of a turkey adenovirus TAdV FJ360746. The consistency of the pyrosequencing technique was appraised, twice repeating the DNA extraction on different days, using different batches of reagents; while the accuracy of the sequences obtained was confirmed by comparison with the results of conventional sequencing.
Nucleotide sequence accession number
FAdV reference strains were previously submitted by the Dr Thierry van den Berg group (Veterinary and Agrochemical Research Centre, Groeselenberg, Brussels, Belgium), which kindly provided us with these viruses (). GenBank accession numbers were assigned to the nucleotide sequences of the FAdV field isolates as follows: 1279/08, HM592267; 147/08, HM592268; 1474/08, HM592269; 132/08, HM592270; 499/08, HM592271; 829/08, HM592272; 457/08, HM592273; 5670, HM592274; 5705, HM592275; 5363, HM592276; 6169, HM592277; 5341, HM592278; 5630, HM592279; 5441, HM592280; 5997, HM592281; 5765, HM592282; 4587, HM592283; 4158, HM592284; 4220, HM592285; 5231, HM592286; 5339, HM592287; 3890, HM592288; 2496, HM592289; 2797, HM592290; 2161, HM592291; 5083, HM592292; and 3636, HM592293 ().
Table 2. Pyrosequencing report of 22 reference strains representing the five FAdV species (A to E) and the 12 different serotypes.
Table 3. Comparison between the BLASTn and the pyrosequence results of the 27 field isolates.
Results
Hexon L1 PCR amplification and sequence analysis
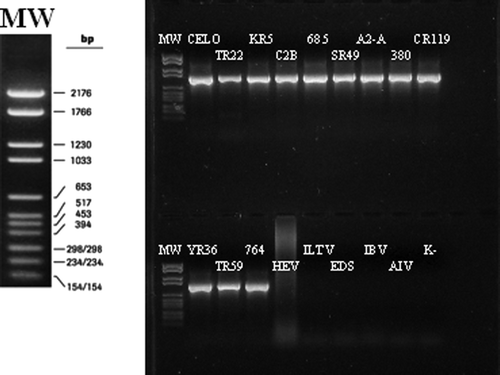
The primer set FadV-Pyro-fw and Hexon B was used to amplify approximately 765 bp of the FAdV hexon gene. All of the samples tested gave a clear amplification product of the expected size, without aspecific bands in a 1.2% agarose gel (). No amplification products were observed for HEV, EDS'76, ILTV, IBV strain M41, AIV subtype H7 and the non-template control (K–).
Figure 2. Agarose gel electrophoresis of the 740 to 765 bp PCR products from single FAdV strains representing FAdV serotypes 1 to 11 and control samples using HEV, EDS'76, ILTV, IBV strain M41, AIV subtype H7 and non-template control (K–). MW, molecular weight.
Sequence analysis of both reference strains and field samples confirmed the hexon L1 amplification and the FAdV strain identity ( and ).
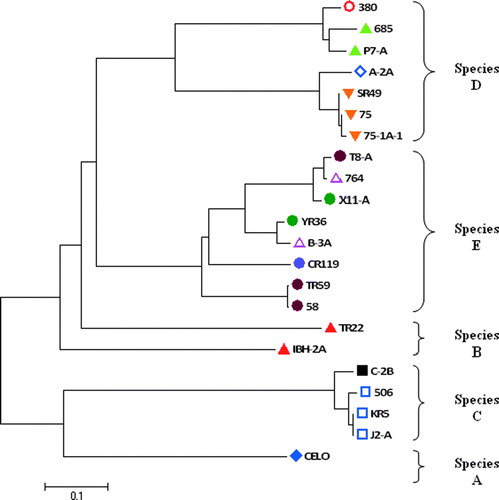
Based on the sequences of reference strains PCR products, a phylogenetic tree was created in order to confirm the clustering of the different species according to their hexon loop sequences ().
Figure 3.
Phylogenetic tree established with the nucleotide sequences of the approximately 765 bp fragment of the reference strains hexon L1 region. The FAdVs are clustered into five species (A to E) and then divided into 12 serotypes according to the ICTV current nomenclature (ICTVdB Management, Citation2006). Serotypes: ![]()
Pyrosequencing analysis of FAdV reference strains
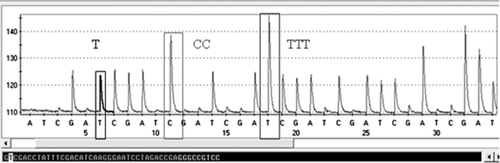
Pyrosequencing analysis of the biotinylated PCR products, using FadV-Pyro-fw as sequencing primer, resulted in good quality pyrograms of up to 30 bp for all the samples tested. shows an example of the resultant pyrogram for a reference strain (506), where peak height is proportional to the amount of pyrophosphate (PPi) and thereby to the number of incorporated nucleotides in the sequence.
Figure 4. Example of pyrogram showing the 30-bp sequence of the hexon L1 of a reference strain (506). The pyrophosphate anion (P2O7 4-, PPi) is released by the hydrolysis of ATP into AMP when a nucleotide is incorporated into a growing DNA strand by the polymerase. Peak height is proportional to the amount of PPi and thereby to the number of incorporated nucleotides.
The related sequences were analysed and compared with a 30-bp hexon sequence library using IdentiFire software (Biotage). According to the reference strain classification (), the pyrosequence results showed a perfect match with the respective species (A to E) for all the viruses with scores ranging from 69.1% to 100% (). In addition it was also possible to distinguish between different serotypes for species D (serotype 11, strain 380; serotype 9, strain A-2A; serotype 3, strains 75, 75-1A-1 and SR49; and serotype 2, strains 685 and P7-A) and species E (serotype 6, strain CR119).
Despite some minor differences in the scores of a few samples, repeating the DNA extraction on different days and using different batches of reagents, the pyrosequencing results were reproducible and viruses were correctly identified, even in the single case where the identity score was as low as 69.1 ().
Owing to sequence homology it was not possible to differentiate between serotypes 4 and 10 belonging to species C and among serotypes 7, 8a and 8b within species E. Indeed, pairwise distance analysis showed that FAdV-10 strain C-2B had 100% sequence homology with FAdV-4 strains 506, J2-A and KR5 in the 30-bp region analysed by pyrosequencing (data not shown).
Trials were also performed, considering the first 50 bp of FAdV hexon L1, in order to increase the number of identifiable serotypes, but no improvement was obtained (results not shown). Furthermore, this approach was more time consuming and there was an increase in the signal background due to the larger amount of pyrosequence reaction waste products.
The situation was the same for strains belonging to serotypes 7, 8a and 8b, but in this case even considering the whole hexon L1 PCR product it would have been impossible to go further than the species detection, because strains classified in different serotypes, by cross-neutralization assay, were too similar in their nucleotide sequences ().
Pyrosequencing analysis of FAdV field samples
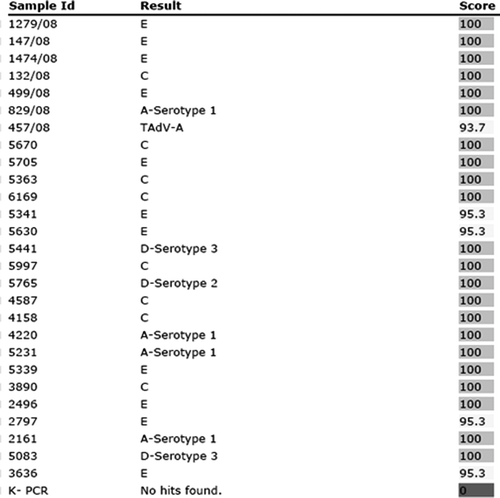
Pyrosequence IdentiFire results for the field isolates were consistent with the corresponding BLASTn results () and had an identification score with the correct species ranging from 93 to 100% as shown in . Of the 27 field samples, isolated both from sick and healthy birds, 11 (41%) belonged to species E, eight (30%) were grouped in species C, while three (11%) and four (15%) belonged to species D and A, respectively. This approach also allowed the identification of two different FAdV serotypes (2 and 3) within one species (D) and a Turkey adenovirus A isolate (sample number 457/08).
Figure 5. IdentiFire report of 27 field samples collected in the north-east of Italy between 2007 and 2008. This approach also allowed the identification of a Turkey adenovirus A isolate (sample number 457/08) of a distinct genogroup. No pyrosequencing reaction was observed for the non-template PCR controls (K–). The results column shows the FAdV species and serotype from the sequence library to which the tested samples shares homology in the 30-bp pyrosequencing region. The degree of homology is reflected by the score, which is given as a percentage.
Discussion
The present study describes the development of a pyrosequencing-based genotyping method and provides a direct comparison between conventional nucleotide sequencing and pyrosequencing analysis for the identification and differentiation of the five FAdV species (A to E).
Pyrosequence results of all samples tested were consistent with the sequence analysis and BLASTn alignment. Based on a 30-bp fragment of the hexon L1 sequence, it was possible to sort all the FAdVs tested into their correct species (A to E), and to directly recognize several serotypes (2, 3, 6, 9 and 11). In addition, it was also possible to identify a Turkey adenovirus A isolate belonging to a different genus (Siadenovirus).
Previous studies have reported the use of techniques such as microtitre virus neutralization tests and PCR in combination with either conventional sequencing or RFLP analysis to identify FAdV species and/or serotypes. Despite the wide use of these techniques there are limitations, including the high cost per analysis, the lengthy processes involved and the need for extensive interpretation (Grimes & King, Citation1977; Erny et al., Citation1995; Steer et al., Citation2009).
Starting from the PCR products, the pyrosequencing analysis required only 2 h, for 96 samples, to obtain the final result; almost one-half the time compared with RFLP analysis or conventional DNA sequencing. Pyrosequencing analysis does not require any restriction enzyme, terminator enzyme or previous purification step; thus resulting in lower cost per analysis when compared with RFLP and DNA sequencing. In fact, an approximate cost calculation including all the reagents necessary for the reaction resulted in an estimate of €3.87 to 6.03 per sample (for 96 or 25 samples analysed/run, respectively), compared with €9.86 per reaction for the classical sequencing method (data not shown). It is also to be noted that, within the same run (same plate), pyrosequencing of different microorganisms with different primers can be performed, increasing the cost-effectiveness and the time effectiveness of the method.
Moreover the results analysis is automatically performed by the software upon completion of the sequencing and is reported in a clear and accurate format, as shown in .
Steer et al. (Citation2009) proposed the use of high-resolution melting curve analysis of the hexon gene region for FAdV classification. However, this analysis was performed only on 12 reference strains, one representative for each serotype, a vaccine strain and just one field sample. Thus its routine application in order to type field samples needs further evaluation.
Based on our sequence analysis of 22 reference strains, it is very difficult to group together certain viruses in the correct serotype because the cross-neutralization classification does not reflect their nucleotide sequences ().
Field samples may possess nucleotide mutation in their sequences that could increase the differences with the reference strains (Meulemans et al., Citation2001). A nucleotide mutation can result in the disappearance of a restriction site in RFLP analysis or a shift in the melting temperature of the high-resolution melting curve analysis, increasing, in both cases, difficulties in interpretation of results. In our study 27 field samples were tested, giving highly accurate nucleotide sequences of 30 bp thanks to the pyrosequencing reaction chemistry (Gharizadeh et al., Citation2006). Moreover, in case of a doubtful result due to few nucleotide mutations, the location and nature of the mismatches are easily identified and reported; thus the correct identification of the virus is still possible by comparisons with other sequences in the databases (e.g. GenBank), which could also be rapidly incorporated in the reference pyrosequence library.
On the strength of these considerations the present study provides a new approach for a rapid differentiation and classification of FAdV species using the pyrosequence analysis that is faster, less expensive and easier to interpret than RFLP or conventional DNA sequencing.
In addition, the study demonstrates that it is possible to obtain excellent pyrosequence results even with biotinylated PCR products larger than the 300 to 500 bp recommended by the manufacturer (Biotage). These results were probably due to the high specificity of the primers and the increased concentration of the streptavidin-coated beads used (5 µl/sample), which enhanced the intensity of the peaks in the pyrogram.
Finally as demonstrated by characterization of FAdVs isolated in different parts of the world, the majority of IBH-associated viruses were grouped in FAdV species E (Erny et al., Citation1991; Saifuddin & Wilks, Citation1991; Ojkic et al., Citation2008) and species D (McFerran et al., Citation1976; Ojkic et al., Citation2008); and these species can be easily and rapidly detected using the pyrosequencing method.
Taken as a whole, the pyrosequencing assay developed here provides a new tool for the detection of FAdV species, including those isolated from field samples, and thus could assist in identifying and rapidly typing different FAdV species for epidemiology tracing and vaccination strategies.
Acknowledgements
The present work was supported by the Italian Ministry of Health (Ricerca Corrente 29/07). The authors wish to thank Dr Thierry van den Berg, Head of Avian Virology & Immunology, Veterinary and Agrochemical Research Centre (VAR), Brussels, for supplying FadV strains 685, 75, 764, 58, 506, YR36, TR22, CR119, KR5, A-2A, X-11A, J2-A, C2B, IBH-2A, B3A, P7-A, 75-1A-1, and T8-A.
References
- Adam , E. , Nasz , I. , Hudecz , F. , Lengyel , A. , Mezo , G. and Dobay , O. 1998 . Characterization of intertype specific epitopes on adenovirus hexon . Archives of Virology , 143 : 1669 – 1682 .
- Alvarado , I.R. , Villegas , P. , El-Attrache , J. , Jensen , E. , Rosales , G. , Perozo , F. and Purvis , L.B. 2007 . Genetic characterization, pathogenicity, and protection studies with an avian adenovirus isolate associated with inclusion body hepatitis . Avian Diseases , 51 : 27 – 32 .
- Athappilly , F.K. , Murali , R. , Rux , J.J. , Cai , Z. and Burnett , R.M. 1994 . The refined crystal structure of hexon, the major coat protein of adenovirus 2, at 2.9 Å resolution . Journal of Molecular Biology , 242 : 430 – 455 .
- Balamurugan , V. and Kataria , J.M. 2004 . The hydropericardium syndrome in poultry—a current scenario . Veterinary Research Communications , 28 : 127 – 148 .
- Christensen , N.H. and Saifuddin , M. 1989 . A primary epidemic of inclusion body hepatitis in broilers . Avian Diseases , 33 : 622 – 630 .
- Crawford-Miksza , L. and Schnurr , D.P. 1996 . Analysis of 15 adenovirus hexon proteins reveals the location and structure of seven hypervariable regions containing serotype-specific residues . Journal of Virology , 70 : 1836 – 1844 .
- Deyde , V.M. and Gubareva , L.V. 2009 . Influenza genome analysis using pyrosequencing method: current applications for a moving target . Expert Review of Molecular Diagnostics , 9 : 493 – 509 .
- Deyde , V.M. , Nguyen , T. , Bright , R.A. , Balish , A. , Shu , B. Lindstrom , S. 2009 . Detection of molecular markers of antiviral resistance in influenza A (H5N1) viruses using a pyrosequencing method . Antimicrobial Agents and Chemotherapy , 53 : 1039 – 1047 .
- Elahi , E. , Pourmand , N. , Chaung , R. , Rofoogaran , A. , Boisver , J. Samimi-Rad , K. 2003 . Determination of hepatitis C virus genotype by pyrosequencing . Journal of Virological Methods , 109 : 171 – 176 .
- Erny , K.M. , Barr , D.A. and Fahey , K.J. 1991 . Molecular characterization of highly virulent fowl adenoviruses associated with outbreaks of inclusion body hepatitis . Avian Pathology , 20 : 597 – 606 .
- Erny , K. , Pallister , J. and Sheppard , M. 1995 . Immunological and molecular comparison of fowl adenovirus serotypes 4 and 10 . Archives of Virology , 140 : 491 – 501 .
- Gharizadeh , B. , Akhras , M. , Nourizad , N. , Ghaderi , M. , Yasuda , K. , Nyrén , P. and Pourmand , N. 2006 . Methodological improvements of pyrosequencing technology . Journal of Biotechnology , 124 : 504 – 511 .
- Grgic , H. , Philippe , C. , Ojkic , D. and Nagy , E. 2006 . Study of vertical transmission of fowl adenoviruses . Canadian Journal of Veterinary Research , 70 : 230 – 233 .
- Grimes , T.M. and King , D.J. 1977 . Serotyping avian adenoviruses by a microneutralization procedure . American Journal of Veterinary Research , 38 : 317 – 321 .
- Grimes , T.M. , King , D.J. , Kleven , S.H. and Fletcher , O.J. 1977 . Involvement of a type-8 avian adenovirus in the etiology of inclusion body hepatitis . Avian Diseases , 21 : 26 – 38 .
- Hess , M. 2000 . Detection and differentiation of avian adenoviruses: a review . Avian Pathology , 29 : 195 – 206 .
- Hess , M. , Raue , R. and Prusas , C. 1999 . Epidemiological studies on fowl adenovirus isolated from cases of infectious hydropericardium . Avian Pathology , 28 : 433 – 439 .
- ICTVdB Management 2006 . 00.001.0.02.001. Fowl adenovirus A C. Büchen-Osmond ICTVdB—The Universal Virus Database (version 4) . New York : Columbia University .
- Jadhao , S.J. , Deepak , J.N. , Kataria , J.M. , Kataria , R.S. , Tiwari , A.K. Somvanshi , R. 2003 . Characterisation of fowl, adenoviruses from chickens affected with infectious hydropericardium during 1994–1998 in India . Indian Journal of Experimental Biology , 41 : 321 – 327 .
- McFerran , J.B. and Connor , T.J. 1977 . Further studies on the classification of fowl adenoviruses . Avian Diseases , 21 : 585 – 595 .
- McFerran , J.B. and Smyth , J.A. 2000 . Avian adenoviruses . Revue Scientifique et Technique (International Office of Epizootics) , 19 : 589 – 601 .
- McFerran , J.B. , McCracken , R.M. , Connor , T.J. and Evans , R.T. 1976 . Isolation of viruses from clinical outbreaks of inclusion body hepatitis . Avian Pathology , 5 : 315 – 324 .
- Meulemans , G. , Boschmans , M. , Berg , T.P. and Decaesstecker , M. 2001 . Polymerase chain reaction combined with restriction enzyme analysis for detection and differentiation of fowl adenoviruses . Avian Pathology , 30 : 655 – 660 .
- Muroga , N. , Taharaguchi , S. , Ohta , H. , Yamazaki , K. and Takase , K. 2006 . Pathogenicity of fowl adenovirus isolated from gizzard erosions to immuno-suppressed chickens . The Journal of Veterinary Medical Science , 68 : 289 – 291 .
- Nakamura , K. , Mase , M. , Yamaguchi , S. , Shibahara , T. and Yuasa , N. 1999 . Pathologic study of specific-pathogen-free chicks and hens inoculated with adenovirus isolated from hydropericardium syndrome . Avian Diseases , 43 : 414 – 423 .
- Nakamura , K. , Tanaka , H. , Mase , M. , Imada , T. and Yamada , M. 2002 . Pancreatic necrosis and ventricular erosion in adenovirus-associated hydropericardium syndrome of broilers . Veterinary Pathology , 39 : 403 – 406 .
- Norby , E. 1969 . The relationship between the soluble antigens and the virion of adenovirus type 3. IV. Immunological complexity of soluble components . Virology , 37 : 565 – 576 .
- Ojkic , D. , Krell , P.J. , Tuboly , T. and Nagy , E. 2008 . Characterization of fowl adenoviruses isolated in Ontario and Quebec, Canada . Canadian Journal of Veterinary Research , 72 : 236 – 241 .
- Ono , M. , Okuda , Y. , Yazawa , S. , Imai , Y. , Shibata , I. , Sato , S. and Okada , K. 2003 . Adenoviral gizzard erosion in commercial broiler chickens . Veterinary Pathology , 40 : 294 – 303 .
- Ono , M. , Okuda , Y. , Yazawa , S. , Shibata , I. , Tanimura , N. Kimura , K. 2001 . Epizootic outbreaks of gizzard erosion associated with adenovirus infection in chickens . Avian Diseases , 45 : 268 – 275 .
- Philippe , C. , Grgic , H. , Ojkic , D. and Nagy , E. 2007 . Serologic monitoring of a broiler breeder flock previously affected by inclusion body hepatitis and testing of the progeny for vertical transmission of fowl adenoviruses . Canadian Journal of Veterinary Research , 71 : 98 – 102 .
- Quince , C. , Lanzen , A. , Curtis , T.P. , Davenport , R.J. , Hall , N. Head , I.M. 2009 . Accurate determination of microbial diversity from 454 pyrosequencing data . Nature Methods , 6 : 639 – 641 .
- Raue , R. and Hess , M. 1998 . Hexon based PCRs combined with restriction enzyme analysis for rapid detection and differentiation of fowl adenoviruses and egg drop syndrome virus . Journal of Virological Methods , 73 : 211 – 217 .
- Roberts , M.M. , White , J.L. , Grutter , M.G. and Burnett , R.M. 1986 . Three-dimensional structure of the adenovirus major coat protein hexon . Science , 232 : 1148 – 1151 .
- Ronaghi , M. and Elahi , E. 2002 . Pyrosequencing for microbial typing . Journal of Chromatography B , 782 : 67 – 72 .
- Saifuddin , M. and Wilks , C.R. 1991 . Pathogenesis of an acute viral hepatitis: inclusion body hepatitis in the chicken . Archives of Virology , 116 : 33 – 43 .
- Sheppard , M. , McCoy , R.J. and Werner , W. 1995 . Genomic mapping and sequence analysis of the fowl adenovirus serotype 10 hexon gene . Journal of General Virology , 76 : 2595 – 2600 .
- Steer , P.A. , Kirkpatrick , N.C. , O'Rourke , D. and Noormohammadi , A.H. 2009 . Classification of fowl adenovirus serotypes by use of high-resolution melting-curve analysis of the hexon gene region . Journal of Clinical Microbiology , 47 : 311 – 321 .
- Swan , D.C. , Limor , J.R. , Duncan , K.L. , Rajeevan , M.S. and Unger , E.R. 2006 . Human papillomavirus type 16 variant assignment by pyrosequencing . Journal of Virological Methods , 136 : 166 – 170 .
- Tamura , K. , Dudley , J. , Nei , M. and Kumar , S. 2007 . MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0 . Molecular Biology and Evolution , 24 : 1596 – 1599 .
- Toogood , C.I. , Crompton , J. and Hay , R.T. 1992 . Antipeptide antisera define neutralizing epitopes on the adenovirus hexon . Journal of General Virology , 73 : 1429 – 1435 .
- Toro , H. , Gonzalez , C. , Cerda , L. , Morales , M.A. , Dooner , P. and Salamero , M. 2002 . Prevention of inclusion body hepatitis/hydropericardium syndrome in progeny chickens by vaccination of breeders with fowl adenovirus and chicken anemia virus . Avian Diseases , 46 : 547 – 554 .
- Toro , H. , Prusas , C. , Raue , R. , Cerda , L. , Geisse , C. , Gonzalez , C. and Hess , M. 1999 . Characterization of fowl adenoviruses from outbreaks of inclusion body hepatitis/hydropericardium syndrome in Chile . Avian Diseases , 43 : 262 – 270 .