Abstract
At the end of 2002 and throughout 2003, there was a severe outbreak of infectious laryngotracheitis (ILT) in an intensive production area of commercial hens in the São Paulo State of Brazil. ILT virus was isolated from 28 flocks, and 21 isolates were genotyped by polymerase chain reaction and restriction fragment length polymorphism (PCR-RFLP) using four genes and eight restriction enzymes, and by partial sequencing of the infected cell protein 4 (ICP4) and thymidine kinase (TK) genes. Three groups resulted from the combinations of PCR-RFLP patterns: 19 field isolates formed Group I, and the remaining two isolates together with the chicken embryo origin (CEO) vaccine strains formed Group II. Group III comprised the tissue-culture origin (TCO) vaccine strain by itself. The PCR-RFLP results agreed with the sequencing results of two ICP4 gene fragments. The ICP4 gene sequence analysis showed that the 19 field isolates classified into Group I by RFLP-PCR were identical among themselves, but were different to the TCO and CEO vaccines. The two Group II isolates could not be distinguished from one of the CEO vaccines. The nucleotide and amino acid sequence analyses discriminated between the Brazilian and non-Brazilian isolates, as well as between the TCO and CEO vaccines. Sequence analysis of the TK gene enabled classification of the field isolates (Group I) as virulent and non-vaccine. This work shows that the severe ILT outbreak was caused by a highly virulent, non-vaccine strain.
Introduction
Infectious laryngotracheitis (ILT) is a highly contagious, upper respiratory disease of chickens that may result in great economic loss because of increased mortality, decreased growth rates and reduced egg production (Guy & García, Citation2008). The aetiologic agent is infectious laryngotracheitis virus (ILTV) or Gallid herpesvirus 1 (GaHV-1), a member of the family Herpesviridae, subfamily Alphaherpesvirus (Roizman, Citation1996). The capacity of ILTV strains to maintain a state of latency in the chickens, in particular within the trigeminal ganglion, is a distinct characteristic of GaHV-1 that makes the disease difficult to control (Williams et al., Citation1992). Two types of ILTV live-attenuated vaccines have been used widely for the control of ILT; the vaccines attenuated by sequential passages in embryonated eggs (chicken embryo origin (CEO)) (Samberg et al., Citation1971); and the vaccine generated after multiple passages in tissue culture (tissue-culture origin (TCO) (Gelenczei & Marty, Citation1964). Recently, ILTV recombinant vaccines have been used in the field (Davison et al., Citation2006). Attenuated vaccines, principally the CEO vaccines, have been isolated from ILT outbreaks from different parts of the world (Graham et al., Citation2000; Kirkpatrick et al., Citation2006; Ojkic et al., Citation2006; Oldoni & García, Citation2007; Neff et al., Citation2008; Oldoni et al., Citation2008) because the attenuated virus can spread from vaccinated bird to non-vaccinated bird, and revert to the virulent form after sequential bird-to-bird passage (Guy et al., Citation1991) or reactivation from latency (Hughes et al., Citation1991). As a consequence, an important aspect of epidemiological studies has included the differentiation between field isolates and vaccine strains (Chang et al., Citation1997; Graham et al., Citation2000; Han & Kim, Citation2001; Creelan et al., Citation2006, Kirkpatrick et al., Citation2006; Ojkic et al., Citation2006; Oldoni & García, Citation2007; Neff et al., Citation2008; Oldoni et al., Citation2008).
Molecular techniques, including conventional and real-time polymerase chain reaction (PCR) assays, have been used efficiently in the diagnosis of ILT (Creelan et al., Citation2006; Callison et al., Citation2007; Chacón & Ferreira, Citation2008). However, the characterization of the type of ILTV that is circulating in the field and/or involved in clinical outbreaks is complicated because of the high antigenic and genetic similarity among the ILT viruses (Guy & García, Citation2008). Initially, epidemiological studies were performed using a restriction fragment length polymorphism (RFLP) analysis of the entire viral genome (Leib et al., Citation1986; Andreasen et al., Citation1990; Keller et al., Citation1992; Keeler et al., Citation1993). This approach was replaced by RFLP of PCR products of multiple genes and genome regions, allowing for the differentiation of ILTV field isolates and vaccine strains. This PCR-RFLP analysis includes the simultaneous amplification of several genes, including the infected cell protein 4 (ICP4), thymidine kinase (TK), glycoprotein G (gG) and glycoprotein E (gE) genes (Chang et al., Citation1997; Graham et al., Citation2000; García & Riblet, Citation2001; Kirkpatrick et al., Citation2006; Ojkic et al., Citation2006; Oldoni & García, Citation2007). Recently, a new approach for the discrimination of ILTV isolates using sequence analysis of the ICP4 gene was performed and validated (Chacón & Ferreira, Citation2009). This assay distinguished between attenuated vaccine strains and field isolates from different geographical regions, and also discriminated between the CEO vaccines of different origin. The specific sequence analysis of IPC4 is accurate and does not require the amplification of several genes nor the virus propagation that is required for PCR-RFLP. In addition, a novel technique—the reverse RFLP assay—was applied to differentiate between the two types of ILTV attenuated live vaccines (Callison et al., Citation2009). On the other hand, sequence analysis of the TK gene has been proposed for determining the virulence of ILTV isolates (Han & Kim, Citation2001).
Before 2002, no severe outbreaks of ILT had been reported in Brazil, and the use of ILT vaccines was not allowed. However, at the end of 2002 and during 2003, an epidemic of severe outbreaks of respiratory disease with high mortality was observed in the commercial laying flocks from the Bastos region of the São Paulo State. Clinical diagnoses suggested that the outbreak was caused by ILTV by the observed clinical signs and lesions, and the diagnosis was then confirmed by PCR, while other respiratory pathogens were ruled out (Chacón et al., Citation2007). During the outbreak, more than 1 million birds died due to intense clinical manifestations, and, as a result, great economic loss was recorded. Importantly, ILTV was detected in young chickens that showed clinical signs. Although the disease was more intensive in farms with poor biosecurity measures, all of the farms from the Bastos region were affected, making necessary the implementation of management procedures and quarantine measures in the region. In addition, the TCO vaccine strain and two CEO vaccines were licensed for vaccination use only in the Bastos region. The exact origin of the outbreak has not been determined, and a characterization of the strains that were involved has not yet been performed. Therefore, the objective of the present study was to genotype the viral isolates that were involved in the severe outbreak by PCR-RFLP of multiple genes and by DNA sequencing of the ICP4 gene.
Materials and Methods
Case history and field samples
In the spring of 2002 and throughout 2003, a significant outbreak of respiratory disease was observed in the Bastos region of the São Paulo State of Brazil. The Bastos region is an intensive poultry production area that includes close to 180 commercial hen farms. All of the farms in the region were affected during the epidemic outbreak, with chickens exhibiting depression, conjunctivitis, gasping, coughing, nasal discharge, marked dyspnoea, expectoration of blood-stained mucous, reduced egg production and an increased mortality rate that ranged from 2 to 20%. The clinical manifestations were more intense in farms with a poor level of biosecurity and in laying hens, but mortality was also observed in birds as young as 11 weeks of age. The farms had intensive vaccination programmes against infectious bronchitis virus and Newcastle disease but no vaccination against ILT. Furthermore, laboratory tests ruled out avian influenza virus. The clinical signs suggested ILT, which was initially confirmed by molecular assays (Chacón et al., Citation2007). Then, trachea, conjunctiva and lung samples from 55 flocks between 11 and 104 weeks old with and without clinical signs were collected during the outbreak by authorities of CDA-SAA of the São Paulo State for ILTV detection and characterization. The three different tissues were pooled from a total of eight birds per flock, were homogenized as 10% (w/v) suspensions in phosphate-buffered saline (pH 7.4), and were then clarified by centrifugation at 3000 x g for 20 min at 4°C. The supernatant was then used for virus propagation and DNA extraction.
Reference strains
One TCO vaccine (LT-Ivax (Samberg strain); Schering Plough Laboratory) and two CEO vaccines (Nobilis ILT (Serva strain); Intervet Laboratory and Laryngo-Vac; Ford Dodge Laboratory) were used as positive controls.
ILTV propagation
The field viruses were propagated in embryonated eggs. Supernatant of homogenized tissues was passed through a 20 µm filter and mixed with 30 µg/ml gentamicin. A suspension of 200 µl was inoculated via the chorioallantoic membrane (CAM) into five 9-day-old embryonated specific-pathogen-free hens’ eggs (Biovet Laboratory). Six days after inoculation, the CAMs were harvested, homogenized and serially passaged four times. After the last passage, the suspension of harvested material (CAM) was submitted to molecular analysis.
Extraction of viral DNA
DNA from field samples, homogenized CAM plaques and commercial vaccine strains was extracted according to Chomczynski (Citation1993) and Chacón & Ferreira (Citation2008). The negative control was phosphate-buffered saline (pH 7.4, 0.01 M). The DNA was dissolved in 30 µl Tris-EDTA (TE) buffer and was stored at –20°C. DNA extracted from the clinical tissue and CAM plaques was used for characterization by DNA sequencing and PCR-RFLP, respectively.
ILTV screening
Samples from the 55 flocks were screened for ILTV with primers and reaction conditions used to amplify a 219-bp fragment corresponding to a region of the ILTV gE gene, as described by Chacón & Ferreira (Citation2008). The ILTVs detected in the screening were submitted to molecular characterization.
PCR-RFLP analysis
DNA extracted from the vaccine strains and 21 field isolates detected in the screening assay was analysed by PCR-RFLP. The field isolates were randomly selected and had been propagated in embryonated eggs. Amplification of the genome regions and genes TK (Han & Kim, Citation2001), gE (García & Riblet, Citation2001), UL47/gG (Oldoni & García, Citation2007) and ICP4 (Chang et al., Citation1997) was performed as described by Chacón & Ferreira (Citation2009), but with some modifications. Ten-microlitres of the PCR products of each of the four genes were digested separately at 37°C for 3 h with 10 µl restriction endonuclease (RE) enzymes. The TK gene fragment was digested with REs HaeIII, Sau96I and NciI; the gE fragment was digested with DdeI; the UL47/gG fragment was digested with NlaIV; and the ICP4 fragment was digested with HaeIII, MspI and HinP1I—all of the digests followed the manufacturer's recommendations (New England Biolabs, Beverly, Massachusetts, USA). After digestion, the restriction DNA fragments were separated on a 0.8% agarose gel and the size of DNA fragments was compared with a molecular size marker (100-bp; Gibco, Groningen, The Netherlands). The combinations of the DNA band patterns that were generated after digestion of the four different genome regions were used to classify the ILTV field isolates and vaccine strains.
PCR of the infected cell protein 4 gene
The 21 field isolates characterized by PCR-RFLP were also analysed by sequencing two ICP4 gene fragments. The DNA that was extracted directly from the clinical tissues from 21 selected farms was submitted to PCR for amplification of two ICP4 gene fragments. The primer pairs ICP4-1F/ICP4-1R and ICP4-2F/ICP4-2R were used for amplification of fragments 688-bp and 635-bp in length, respectively, as described by Chacón & Ferreira (Citation2009). The PCR products were separated by electrophoresis on a 1.5% w/v agarose gel and were stained with an ethidium bromide solution (0.5 µg/ml).
PCR of the thymidine kinase gene
DNA from the attenuated vaccine strains and from eight field isolates was subjected to a nested PCR assay to amplify a fragment of the TK gene. The primers and reaction conditions were as published by Han & Kim (Citation2001) and were used to amplify a 649-bp fragment.
Sequencing and sequence analysis
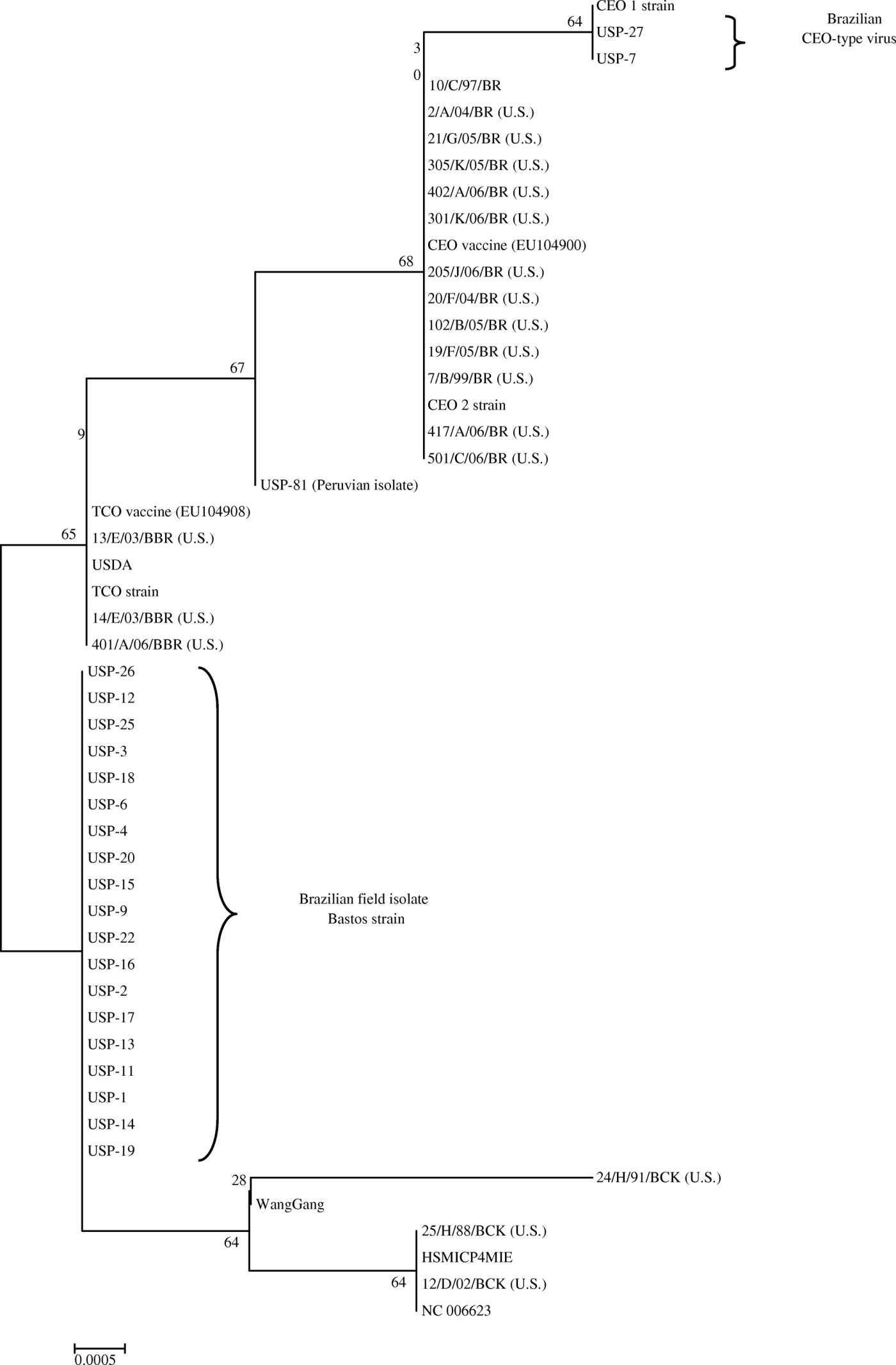
The 688-bp and 635-bp (ICP4 gene) PCR products and the 649-bp (TK gene) PCR product from the ILTV-positive samples were submitted to DNA sequencing. The products were purified using a GFX™ PCR DNA and Gel Band Purification Kit (GE Healthcare, Piscataway, New Jersey, USA) as described by the manufacturer. Each purified product was then sequenced in the forward and reverse directions according to the instructions of the BigDye Terminator v3.1 Cycle Sequencing kit (Applied Biosystems, Foster City, California, USA). Sequencing reactions were run in an ABI PRISM 3730 Genetic Analyzer (Applied Biosystems). The data sequences were assembled and analysed using the BioEdit version 7.0.5 package. The nucleotide sequences were determined for both strands and were checked twice. The nucleotide and deduced amino acid sequences of the two ICP4 gene fragments were aligned and compared using CLUSTALX software. Phylogenetic trees were generated by the neighbour-joining methods with 1000 bootstrap replicates using the MEGA software program version 3.1 (Kumar et al., Citation2001).
Nucleotide sequence accession numbers
For sequence analysis, the published sequences in Genbank were used: USDA (accession number EU104909), HSMICP4MIE (accession number L32139.1), (accession number NC_006623), Wang Gang (accession number DQ995291), CEO vaccine (accession number EU104900), TCO vaccine (accession number EU104908), 25/H/88/BCK (accession number EU104899), 19/F/05/BR (accession number EU104901), 21/G/05/BR (accession number EU104902), 301/K/06/BR (accession number EU104903), 10/C/97/BR (accession number EU104904), 401/A/06/BBR (accession number EU104905), 14/E/03/BBR (accession number EU104906), 13/E/03/BBR (accession number EU104907), 24/H/91/BCK (accession number EU104910), 12/D/02/BCK (accession number EU104911), 305/K/05/BR (accession number EU104913), 20/F/04/BR (accession number EU104915), 205/J/06/BR (accession number EU104916), 402/A/06/BR (accession number EU104917), 102/B/05/BR (accession number EU104918), 7/B/99/BR (accession number EU104919) and 501/C/06/BR (accession number EU104920) for ICP4 gene; 216 (accession number L36139), U.S. 632 (accession number S83714), Beijing E2 (accession number AF435453), XY (accession number DQ522946), Samberg (accession number DQ522947), GG (accession number DQ522949), Lacp (accession number D00565) and Yantai (accession number AY741134) for TK gene.
Results
PCR-RFLP analysis
Of the 55 farms tested, ILTV was detected in 25 cases (in 13 farms, clinical signs were observed during the collection of samples). Twenty-one field isolates were then randomly selected and characterized by PCR-RFLP. One TCO vaccine and two CEO vaccine strains were included in the study for molecular comparison. The results of the PCR-RFLP analysis of TK, UL47/gG and ICP4 (using the RE HaeIII) were identical, and it was possible to classify the 21 field isolates and three attenuated vaccine strains into two groups. Nineteen field samples were categorized into Group I, while the remaining two field samples (USP-7 and USP-27) and the TCO and CEO vaccines formed Group II. The PCR-RFLP analysis of the E gene generated two groups; Group I included all of the field isolates and the two CEO vaccines, while Group II included only the TCO vaccine. Three different RFLP patterns were generated by digestion of the 4.9-kb ICP4 gene PCR product using MspI and HinP1I (). Similar to the results described above, 19 field isolates were categorized into Group I, the USP-7 and USP-27 isolates as well as the two CEO vaccines that were included in this study were categorized into Group II, and the TCO vaccine was categorized into Group III. The USP-7 and USP-27 isolates could not be differentiated from the CEO vaccines analysed in the present study after multiple PCR-RFLP analyses of the four PCR products ().
Figure 1. HaeIII (1a), MspI (1b) and HinP1I (1c) digests of the 4.9 kb PCR product from the ICP4 gene from the vaccine strains and representative Brazilian field isolates of ILTV. Arrows indicate differences in the RFLP patterns. Lane 1, molecular size marker (100 bp; Invitrogen®); lane 2, USP-1; lane 3, USP-7; lane 4, TCO strain; lane 5, CEO 1 strain; lane 6, CEO 2 strain.
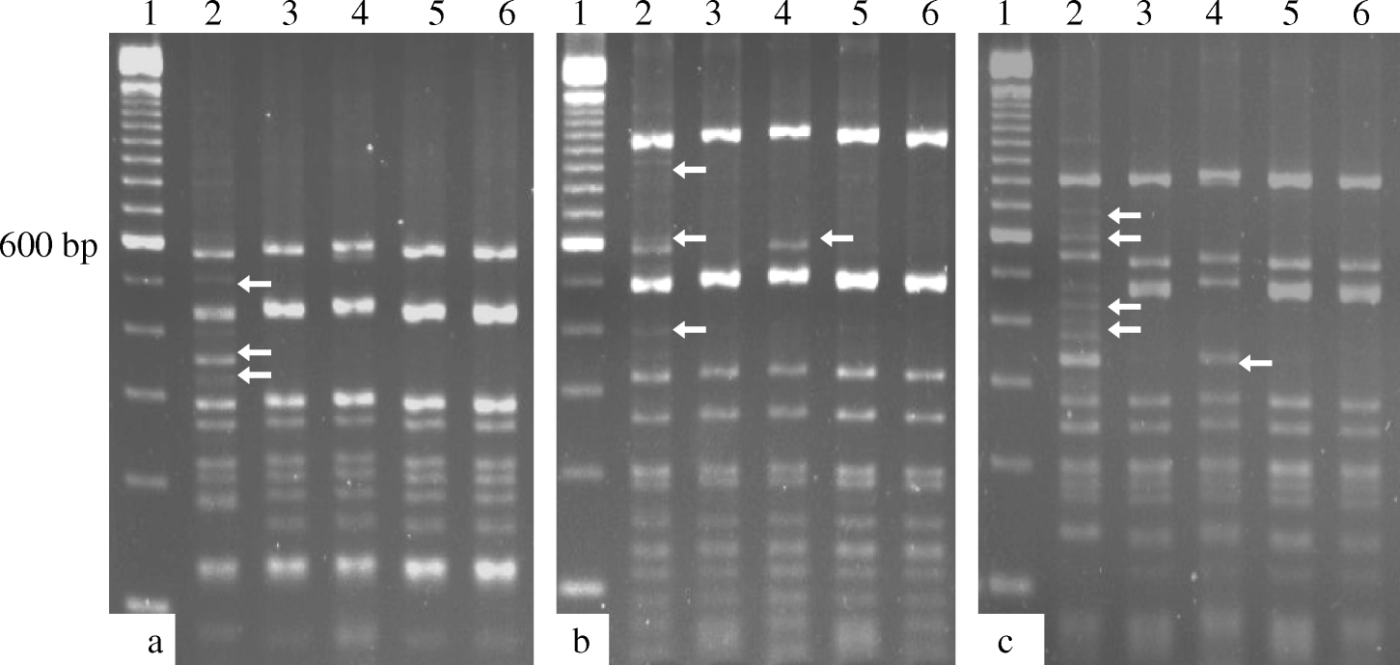
Table 1. Epidemiological information, PCR-RFLP analysis, virulence according to TK gene sequence analysis and results of the sequence analysis of two ICP4 gene fragments from the ILTV field isolates and vaccine strains.
PCR and sequence analysis of the ICP4 gene
The same field isolates characterized by PCR-RFLP were also analysed by sequencing of the ICP4 gene. PCR was used to amplify 688-bp and 635-bp fragments using the primer pairs ICP4-1F/ICP41R and ICP4-2F/ICP4-2R, respectively. Both of the reactions amplified DNA from the ILTV field isolates and vaccine strains, whereas no products were found in the negative control reactions.
Sequence analysis of the 688-bp fragments showed 100% nucleotide identity among all of the Brazilian field isolates, with the exception of isolates USP-7 and USP-27. The 19 field isolates had 99.8%, 97.4 to 97.5%, and 97.5 to 99.8% similarity with the TCO, CEO vaccines, and non-Brazilian field isolates, respectively (data not shown). The USP-7 and USP-27 Brazilian isolates had a high similarity to the CEO vaccine sequences, with 100% identity with one of them (CEO-1). Deduced amino acid sequence analysis differentiated between the TCO and CEO vaccines. The TCO vaccine and the 19 Brazilian field isolates differed from the CEO vaccines by four additional amino acids (two alanines, glycine and aspartic acid). Finally, the CEO-1 and CEO-2 vaccines were differentiated from one another by nucleotide and amino acid sequences analysis ().
Table 2. Nucleotide sequence comparisons of the two ICP4 gene fragments (from nucleotides 181 to 869 and from nucleotides 3804 to 4440 of the gene) from the Brazilian field isolates, vaccine strains and other published non-Brazilian ILTV field isolates.
Similar results were obtained after analysis of the 635-bp fragments of the ICP4 gene. The Brazilian field isolates had 100% nucleotide identity, with the exception of isolates USP-7 and USP-27. The 19 identical field isolates had 99.1%, 99.3 to 99.4%, and 95.7 to 99.8% nucleotide identity when compared with the TCO vaccine, the CEO vaccine strains, and the non-Brazilian field isolates, respectively (data not shown). Similar to results with the 688-bp fragment, the 635-bp fragments from the USP-7 and USP-27 isolates were also indistinguishable from CEO-1. The TCO and CEO vaccines could be differentiated from one another by nucleotide and amino acid sequence analysis. Additionally, the two CEO vaccines were differentiated from one another by nucleotide sequence analysis ().
The Brazilian field isolates (except USP-7 and USP-27) were differentiated from attenuated vaccine strains by phylogenetic trees constructed from the two ICP4 gene fragments (). The sequences obtained in this study were published in the Genbank database with the following accession numbers: LT-IVAX (FJ477349), Laryngo-Vac (FJ477350), Nobilis ILT (FJ477351), USP-01 (FJ477352), USP-02 (FJ477353), USP-03 (FJ477354), USP-04 (FJ477355), USP-06 (FJ477356), USP-07 (GQ499345), USP-09 (FJ477357), USP-11 (GQ499346), USP-12 (FJ477358), USP-13 (FJ477359), USP-14 (GQ499347), USP-15 (GQ499348), USP-16 (GQ499349), USP-17 (GQ499350), USP-18 (GQ499351), USP-19 (FJ477360), USP-20 (FJ477361), USP-22 (FJ477362), USP-25 (FJ477363), USP-26 (FJ477364) and USP-27 (FJ477365).
Agreement between PCR-RFLP and sequence analysis
Similar results were obtained using PCR-RFLP and sequence analysis. PCR-RFLP analysis and sequencing of the ICP4 gene differentiated the 19 Brazilian isolates from the attenuated vaccines. However, isolates USP-7 and USP-27 were not differentiated from the CEO vaccines by either method. In fact, the nucleotide sequences from these field isolates were identical to sequences of CEO-1 vaccine. The two types of attenuated vaccines (TCO and CEO) were discriminated from one another by both of the approaches. However, only DNA sequencing distinguished between the two CEO vaccines (). PCR-RFLP analysis and DNA sequencing of the ICP4 gene had similar results, in that the DNA sequencing provided more discrimination between the analysed ILTV isolates and strains.
PCR and sequence analysis of the TK gene
The nucleotide sequences from all the Brazilian isolates were 100% identical, with the exception of isolates USP-7 and USP-27. The Brazilian isolates had 99.5%, 99.6%, and 99 to 99.6% similarity with TCO, CEO and non-Brazilian field strains, respectively. On the other hand, isolates USP-7 and USP-27 were 99.8% and 100% similar to the TCO and CEO vaccines, respectively (data not shown). The deduced amino acid sequence analysis showed that the vaccine strains had a threonine in position 252 of the gene, whereas the Brazilian field isolates (except USP-7 and USP-27) had a methionine in the same position. The sequences obtained in the present study were published in the Genbank database with the following accession numbers: LT-IVAX (FJ444832), Laryngo-Vac (FJ444830), Nobilis ILT (FJ444830), USP-01 (FJ444833), USP-02 (FJ444834), USP-06 (FJ444835), USP-07 (GQ499341), USP-09 (FJ444836), USP-22 (DQ786400), USP-26 (FJ444837), and USP-27 (DQ786401).
Discussion
The present study investigated the origins of the ILTV strains involved in the first severe outbreaks of ILT in Brazil, which affected commercial layer flocks from an intensive poultry production region of the São Paulo State. PCR-RFLP and sequence analysis were used to genotype the viruses involved in the outbreak and the results obtained were compared.
The PCR-RFLP analysis of the TK and UL47/gG genes allowed for differentiation between the field isolates and vaccine strains only. Analysis of the gE gene distinguished the TCO vaccine from the field isolates and the CEO vaccines. In all of the cases, the USP-7 and USP-27 isolates were classified as vaccine strains. On the other hand, PCR-RFLP analysis of the ICP4 gene showed greater discriminatory power. Nineteen field isolates were differentiated from the vaccine strains using the REs MspI and HinP1I. In addition, the TCO and CEO vaccine-type strains were distinguished from one another, while the USP-7 and USP-27 isolates had the same RFLP pattern as the CEO vaccines. These results are in concordance with published studies, which showed the usefulness of the ICP4 gene for the genotyping of ILT isolates (Chang et al., 1997; Graham et al., Citation2000; Creelan et al., Citation2006; Kirkpatrick et al., Citation2006; Ojkic et al., Citation2006; Oldoni & García, Citation2007; Oldoni et al., Citation2008). These results show that, during the severe outbreak in the Bastos region of Brazil, two CEO vaccine-type ILTVs and a wild-type virus were circulating in the region simultaneously.
The PCR-RFLP approach could determine that the clinical outbreak was caused by a wild-type virus. However, it was first necessary to propagate the virus from the field samples by four passages in embryonated eggs. Other authors have also mentioned that it is necessary to propagate the virus in order to amplify fragments larger than 2 kb (Oldoni & Garcia, 2007; Neff et al., Citation2008; Chacón & Ferreira, Citation2009). In other epidemiological studies, it was also necessary to amplify several genes or genome regions simultaneously, and to use several REs to discriminate efficiently between field isolates and vaccine strains. We have published a new approach for the differentiation of field isolates, TCO and CEO vaccines from one another (Chacón & Ferreira, Citation2009). This approach included the sequencing of two ICP4 gene fragments, and had great discriminatory power. The sequence analysis results of both of the fragments were similar, indicating that either fragment can be used for the discrimination of ILTV strains. In that study, sequence analysis permitted the differentiation of field isolates from different geographic regions, TCO and CEO vaccine strains. Additionally, that approach distinguished between CEO vaccines from different origins. Therefore, 21 ILTV isolates that were collected from the Bastos region during the epidemic outbreak were submitted to the described approach.
Agreement was observed between the single-gene sequence analysis and the multiple-gene PCR-RFLP analysis. In fact, the sequence analysis had great discriminatory power because it could distinguish between the two CEO vaccines. The available gene sequences have shown that ILTV is genetically highly conserved (Guy & García, Citation2008). This molecular characteristic has made it difficult to use DNA sequencing for differentiation of the ILTV strains. Furthermore, DNA sequencing is more rapid and practical than PCR-RFLP because it does not require virus propagation or the amplification of several genes or genome regions and nucleotide sequences can be easily reproduced and published for comparison with other viruses that have been isolated from different geographic regions.
From the 21 ILTVs characterized in the present study, 19 were categorized as field or wild isolates, while the two remaining viruses could not be differentiated from the CEO vaccines. The 19 field isolates were shown to be identical by PCR-RFLP and sequencing analysis, suggesting that they all belonged to the same strain, referred to in this work as the Bastos strain. Moreover, the USP-7 and USP-27 isolates were CEO vaccine-type viruses and were indistinguishable from one CEO vaccine by sequence analysis of two ICP4 gene fragments. The origin of the vaccine-type isolates is unknown because the use of ILT vaccines was not authorized until the time of the outbreak in Brazil. We speculate that the vaccine strain might have been introduced in Brazil through other live vaccines or introduced illegally. CEO vaccines were isolated from the ILT outbreaks, suggesting that they could have caused clinical disease by reversion to virulence after serial passages in susceptible birds or by reactivation from latency (Guy et al., Citation1991; Hughes et al., Citation1991; Oldoni & García, Citation2007; Neff et al., Citation2008; Oldoni et al., Citation2008). Nevertheless, the large number of wild isolates detected in the present study indicates that the outbreak originated from a virulent wild-type strain.
Based on the results obtained from both of the approaches used in the present study, we can conclude that the ILT outbreak in the Bastos region was caused by one wild-type strain (Bastos strain). This outbreak was characterized by a high incidence of ILTV and low frequency of other respiratory pathogens (Chacón et al., Citation2007). These results demonstrated that no co-infections took place during the outbreak, and that a highly virulent ILTV was the only pathogen responsible for the respiratory outbreak. According to the classification proposed by Han & Kim (Citation2001), six isolates of Bastos strain were characterized as high-virulence virus. Although additional molecular studies are necessary to find all of the genes related to virulence, the severe clinical signs and lesions observed in the field were in agreement with the TK gene sequence analysis and suggest that the Bastos strain is a highly virulent strain. The authors indicate that strains of low virulence have threonine at position 252 in the TK gene sequence, whereas virulent strains have methionine at this position.
The origin of the Bastos strain remains unknown. Kirkpatrick et al. (Citation2006), Ojkic et al. (Citation2006)and Neff et al. (Citation2008) could not differentiate between ILTV isolates from commercial poultry and from backyard flocks. These findings suggest that non-commercial poultry is an important source of infection for commercial flocks because viral latency is favoured in backyard flocks. Recently, a disease survey in backyard chicken flocks from Costa Rica showed that there was a high prevalence of ILTV antibodies (72%) in healthy and ill birds. None of these birds had been vaccinated against Sick (Hernandez-Divers et al., Citation2008). Further epidemiological studies that include Brazilian non-commercial flocks are necessary to determine the participation of these birds in spreading ILTV.
Due to the severe consequences of the epidemic outbreak, management procedures and quarantine measures in the region were implemented and the TCO vaccine strain and the two CEO vaccines included in the present study were licensed for use only in the Bastos region. As a consequence, molecular epidemiology studies will be necessary to evaluate the measures that were implemented to control ILT. The use of ICP4 gene sequence analysis will be very helpful in monitoring this region and neighbouring regions in order to detect elimination or persistence of the wild strain. Importantly, the method can be used to detect the dissemination of the Bastos and/or vaccine strains to non-vaccinated regions.
The results from the present study clearly demonstrate that the respiratory outbreak in the Bastos region was caused by a highly virulent non-vaccine strain. In addition, two CEO vaccine-type ILTVs were detected. The genotyping of viruses was performed with PCR-RFLP analysis of four genes and with eight RE and ICP4 gene sequencing. The sequencing results were in agreement with those obtained by PCR-RFLP. This study shows that sequence analysis of the ICP4 gene can be used efficiently for the discrimination of field strains and different types of attenuated vaccine strains.
Acknowledgements
The authors would like to thank FAPESP (Fundação de Amparo à Pesquisa do Estado de São Paulo, grant number 05/56165-1) for financial support. They also thank the Biovet, Fort Dodge and Merial Laboratories (Brazil) for the provision of embryonated eggs, and thank CDA–SAA, São Paulo for the field sample and vaccine strains they sent.
References
- Andreasen , J.R. , Glisson , J.R. and Villegas , P. 1990 . Differentiation of vaccine strains and Georgia field isolates of infectious laryngotracheitis virus by their restriction endonuclease fragment patterns . Avian Diseases , 34 : 646 – 656 .
- Callison , S.A. , Riblet , S.M. , Oldoni , I. , Sun , S. , Zavala , G. Williams , S. 2007 . Development and validation of a real-time Taqman® assay for the detection and quantitation of infectious laryngotracheitis virus in poultry . Journal of Virological Methods , 139 : 31 – 38 .
- Callison , S.A. , Riblet , S.M. , Rodriguez-Avila , A. and García , M. 2009 . Reverse restriction fragment length polymorphism (RRFLP): a novel technique for genotyping infectious laryngotracheitis virus (ILTV) live attenuated vaccines . Journal of Virological Methods , 160 : 119 – 124 .
- Chacón , J.L. and Ferreira , A.J.P. 2008 . Development and validation of nested-PCR for the diagnosis of clinical infectious laryngotracheitis . Journal of Virological Methods , 151 : 188 – 193 .
- Chacón , J.L. and Ferreira , A.J.P. 2009 . Differentiation of field isolates and vaccine strains of infectious laryngotracheitis virus by DNA sequencing . Vaccine , 27 : 6731 – 6738 .
- Chacón , J.L. , Brandão , P.E. , Villarreal , L. , Gama , N. and Ferreira , A.J.P. 2007 . Survey of infectious laryngotracheitis outbreak in chicken layer hens and differential diagnosis with other respiratory pathogens . Brazilian Journal of Poultry Science , 9 : 61 – 67 .
- Chang , P.C. , Lee , Y.L. , Schien , J.H. and Shieh , H.K. 1997 . Rapid differentiation of vaccine strains and field isolates of infectious laryngotracheitis virus by restriction fragment length polymorphism of PCR products . Journal of Virological Methods , 66 : 179 – 186 .
- Chomczynski , P.A. 1993 . A reagent for the single-step simultaneous isolation of RNA, DNA and protein from the cell and tissues samples . Biotechniques , 15 : 532 – 537 .
- Creelan , J.L. , Calvert , V.M. , Graham , D.A. and McCullough , S.J. 2006 . Rapid detection and characterization from field cases of infectious laryngotracheitis virus by real-time polymerase chain reaction and restriction fragment length polymorphism . Avian Pathology , 35 : 173 – 179 .
- Davison , S. , Gingerich , E.N. , Casavant , S. and Eckroade , R. 2006 . Evaluation of the efficacy of a live fowlpox-vectores infectious laryngotracheitis/avian encephalomyelitis vaccine against ILT viral challenge . Avian Diseases , 50 : 50 – 54 .
- García , M. and Riblet , S.M. 2001 . Characterization of infectious laryngotracheitis virus isolates: demonstration of viral subpopulations within vaccine preparations . Avian Diseases , 45 : 558 – 566 .
- Gelenczei , E.F. and Marty , E.W. 1964 . Studies on a tissue-culture modified infectious laryngotracheitis virus . Avian Diseases , 8 : 105 – 122 .
- Graham , D.A. , McLaren , I.E. , Calvert , V. , Torrens , D. and Meehan , B.M. 2000 . RFLP analysis of recent Northern Ireland isolates of infectious laryngotracheitis virus: comparison with vaccine virus and field isolates from England, Scotland and the Republic of Ireland . Avian Pathology , 29 : 57 – 62 .
- Guy J.S. García M. 2008 Laryngotracheitis In Y.M. Saif J.R. Glisson A.M. Fadly L.R. Mcdougald L.K. Nolan D.E. Swayne Diseases of Poultry , 12th edn 137 152 Ames Iowa State Press
- Guy , J.S. , Barnes , H.J. and Smith , L. 1991 . Increased virulence of modified-live infectious laryngotracheitis vaccine virus following bird-to-bird passage . Avian Diseases , 35 : 348 – 355 .
- Han , M.G. and Kim , S.J. 2001 . Analysis of Korean strains of infectious laryngotracheitis virus by nucleotide sequences and restriction fragment length polymorphism . Veterinary Microbiology , 83 : 321 – 331 .
- Hernandez-Divers , S.M. , Villegas , P. , Jimenez , C. , Hernandez-Divers , S.J. , García , M. Riblet , S.M. 2008 . Backyard chicken flocks pose a disease risk for neotropic birds in Costa Rica . Avian Diseases , 52 : 558 – 566 .
- Hughes , C.S. , Williams , R. , Gaskell , A. , Jordan , R.M. , Brandbury , F.T. , Bennett , J.M. and Jones , R.C. 1991 . Latency and reactivation of infectious laryngotracheitis vaccine virus . Archives of Virology , 121 : 213 – 218 .
- Keeler , C.L. , Hazel , J.W. , Hasting , J.E. and Rosenberger , J.K. 1993 . Restriction endonuclease analysis of Delmarva field isolates of infectious laryngotracheitis virus . Avian Diseases , 37 : 418 – 426 .
- Keller , C.L. , Benson , C.E. , Davison , S. and Eckroade , R.J. 1992 . Differences among restriction endonuclease DNA finger of Pennsylvania field isolates, vaccines strains and challenge strains of infectious laryngotracheitis virus . Avian Diseases , 36 : 575 – 581 .
- Kirkpatrick , N. , Mahmoudian , A. , O'Rourke , D. and Noormohammadi , A.H. 2006 . Differentiation of infectious laryngotracheitis virus isolates by restriction fragment length polymorphic analysis of polymerase chain reaction products amplified from multiple genes . Avian Diseases , 50 : 28 – 34 .
- Kumar , S. , Tamura , I.T. , Jakobsen , I.B. and Nei , M. 2001 . MEGA 2: molecular evolutionary analysis software . Bioinformatics , 17 : 1244 – 1249 .
- Leib , D.A. , Bradbury , J.M. , Gaskell , R.M. , Hughes , C.S. and Jones , R.C. 1986 . Restriction endonuclease patterns of some European and American isolates of avian infectious laryngotracheitis virus . Avian Diseases , 30 : 835 – 837 .
- Neff , C. , Sudler , C. and Hoop , R.K. 2008 . Characterization of Western European field isolates and vaccine strains of avian infectious laryngotracheitis virus by restriction fragment length polymorphism and sequence analysis . Avian Diseases , 52 : 278 – 283 .
- Ojkic , D. , Swinton , J. , Vallieres , M. , Martin , E. , Sharipo , J. , Sanei , B. and Binnington , B. 2006 . Characterization of field isolates of infectious laryngotracheitis virus from Ontario . Avian Pathology , 35 : 286 – 292 .
- Oldoni , I. and García , M. 2007 . Characterization of infectious laryngotracheitis virus isolates from the United States by polymerase chain reaction and restriction fragment length polymorphism of multiple genome regions . Avian Pathology , 36 : 167 – 176 .
- Oldoni , I. , Rodriguez-Avila , A. , Riblet , S. and García , M. 2008 . Characterization of infectious laryngotracheitis virus (ILTV) isolates from commercial poultry by polymerase chain reaction and restriction fragment length polymorphism (PCR-RFLP) . Avian Diseases , 52 : 59 – 63 .
- Roizman , B. 1996 . “ Herpesviridae ” . In Virology , 3rd edn , Edited by: Fields , B.N. , Knipe , D.M. and Howley , P.M. 2221 – 2230 . Philadelphia , PA : Lippincott-Raven .
- Samberg , Y. , Cuperstein , E. , Bendheim , U. and Aronovici , I. 1971 . The development of a vaccine against avian infectious laryngotracheitis. IV. Immunization of chickens with a modified laryngotracheitis vaccine in the drinking water . Avian Diseases , 15 : 413 – 417 .
- Williams , R.A. , Bennett , M. , Bradbury , J.M. , Gaskell , R.M. , Jones , R.C. and Jordan , F.T.W. 1992 . Demonstration of sites of latency of infectious laryngotracheitis virus using the polymerase chain reaction . Journal of General Virology , 73 : 2415 – 2420 .