Abstract
The present study was undertaken to detect and characterize Newcastle disease virus (NDV) in raptors. Cloacal and oropharyngeal swab samples were collected from 60 casualty raptors during January to March 2009 in Minnesota. Inoculation of all these samples (n=120) in 9-day-old embryonated hens’ eggs resulted in isolation of haemagglutinating viruses in three samples from two bald eagles and one great horned owl. These three haemagglutinating viruses were confirmed as NDV by reverse transcription-polymerase chain reaction (RT-PCR) using fusion gene-specific primers, and were negative for avian influenza virus by RT-PCR. Further characterization revealed that all three possessed 112GKQGRL117 at the fusion gene cleavage site, indicating that they were lentogenic strains. Phylogenetic analysis revealed that all three isolates clustered with published class II genotype II NDVs. The nucleotide sequence homology of the three NDV isolates among themselves was 98.4 to 99.6% and the sequence homology with lentogenic strains from wild birds used for comparison varied between 94.5 and 100%. Detection of NDV strains from raptors merits further epidemiological studies to determine the prevalence of different NDV strains in raptors and their impact in relation to transmission to domestic poultry.
Introduction
Newcastle disease virus (NDV), also called avian paramyxovirus type-1, belongs to the genus Avulavirus in the family Paramyxoviridae. This virus exists in two distinct classes (class I and class II) within a single serotype (Czegledi et al., Citation2006). Each class has been further divided into different genotypes; genotypes 1 to 9 in class I, and genotypes I to IX in class II. Based on the level of pathogenicity in chickens, NDVs have been categorized into lentogenic, mesogenic, and velogenic strains (Alexander, Citation2000). Lentogenic strains are the least virulent, mesogenic strains are moderately virulent, and velogenic strains are the most virulent. Both mesogenic and velogenic strains are classified as virulent strains. The virulent strains can cause high morbidity and mortality in commercial poultry, leading to considerable economic losses. In addition, control measures including vaccination are very costly (Alexander, Citation2003). During 2002 and 2003, an outbreak of exotic Newcastle disease in backyard fowl and commercial poultry in the US resulted in the destruction of approximately 3.9 million birds and cost $188 million to control the disease (Wise et al., Citation2004). To avert such losses, continuous surveillance for NDV in different avian species is needed.
NDV can infect a great variety of avian species and has been isolated and characterized from domestic poultry (Marin et al., Citation1996; Jorgensen et al., Citation2004; Pedersen et al., Citation2004; Seal et al., Citation2005) and wild bird populations (Takakuwa et al., Citation1998; Huovilainen et al., Citation2001; Jorgensen et al., Citation2004; Kim et al., Citation2007; Dormitorio et al., Citation2009; Jindal et al., Citation2009; Zhu et al., Citation2010). However, there are only a few reports on the detection and characterization of NDVs from raptors (Keymer & Dawson, Citation1971; Chu et al., Citation1976; Kou et al., Citation1999; Manvell et al., Citation2000; Lublin et al., Citation2001; Schettler et al., Citation2003; Choi et al., Citation2008). A few studies reveal the presence of antibodies to NDV in raptors. Kocan et al. (Citation1977) reported anti-NDV antibody in a red-tailed hawk. Schettler et al. (Citation2001) reported that 2% of the diurnal birds of prey had antibodies to NDV. Hofle et al. (Citation2002) detected NDV antibodies in about 17% of captive and free-living birds. Raptors, especially those that co-mingle with migratory birds during migration, may help disseminate NDV infection from one location to the other. Thus, it is important to study the NDVs circulating in raptor populations, which may elucidate the epidemiology of NDV. The present study was conducted to isolate and characterize NDVs from different species of raptors.
Materials and Methods
Collection of samples
From January to March 2009, under a National Institutes of Health project on avian influenza surveillance, cloacal and oropharyngeal (OP) swabs were collected from 60 casualty raptors admitted to rehabilitation centres in the Upper Midwest, USA, including The Raptor Center at the University of Minnesota. The species sampled were: bald eagle (Haliaeetus leucocephalus, n = 30), great horned owl (Bubo virginianus, n = 11), Cooper's hawk (Accipiter cooperi, n = 10), turkey vulture (Carthartes aura, n = 5), and black vulture (Coragyps atratus, n = 4). The swab samples were collected in 1.0 ml brain heart infusion broth containing antibiotics (penicillin 500 IU/ml, streptomycin 500 µg/ml, neomycin 0.15 mg/ml, fungizone 1.5 µg/ml, and gentamicin 50 µg/ml). The samples were collected immediately on admission of raptors to the rehabilitation centres and only then were the incoming birds mixed with the resident bird population in the rehabilitation centre. After collection, the samples were transported to the laboratory on ice and were stored in the laboratory at –80°C until tested.
Virus isolation
Tubes containing cloacal or OP swabs were vortexed to ensure thorough mixing. The broth was expressed from the swabs before discarding them. The expressed fluid from both cloacal and OP samples was inoculated into two 9-day-old embryonated specific-pathogen-free fowl eggs via the allantoic route using 200 µl sample per egg. The inoculated eggs were incubated at 37°C and candled daily for 4 days. Embryonic death within 24 h of inoculation was considered non-specific, and such eggs were discarded. Eggs showing embryonic death after 24 h and up to day 4 were chilled. Eggs that were viable after 4 days of incubation were also chilled. On the fifth day, allantoic fluid from each of the inoculated eggs was harvested and assayed for haemagglutination (HA) using 0.5% turkey red blood cells. The HA-negative samples were passaged once more in specific-pathogen-free embryonated fowl eggs. The HA titre of the allantoic fluid after the second passage was again determined. All HA-positive allantoic fluids were tested further by reverse transcription-polymerase chain reaction (RT-PCR) for NDV.
Testing of allantoic fluid for NDV by RT-PCR
Total RNA was extracted from the HA-positive allantoic fluid and from a known avian paramyxovirus type-1 obtained from the repository of the Minnesota Veterinary Diagnostic Laboratory (Saint Paul, Minnesota, USA). The QIAamp Viral RNA extraction kit (Qiagen, Valencia, California, USA) was used for total RNA extraction. Extracted RNA was tested for the presence of NDV by RT-PCR using primers (forward, 5′-GCAGCTGCAGGGATTGTGGT-3′ and reverse, 5′-TCTTTGAGCAGGAGGATGTTG-3′) specific to the fusion (F) gene (Nanthakumar et al., Citation2000). Amplification for the detection of NDV was carried out using OneStep RT-PCR kit (Qiagen). The amplified PCR products were gel electrophoresed and a band of 356 bp was observed in positive cases. All HA-positive allantoic fluids were also tested for avian influenza virus (AIV) by RT-PCR using the method of Chan et al. (Citation2006); this method amplifies the matrix gene, a highly conserved AIV gene. A known AIV obtained from the repository of the Minnesota Veterinary Diagnostic Laboratory was used as a positive control. No HA-positive allantoic fluid was found positive for AIV.
Sequencing
The NDV-positive PCR products were purified using a PCR purification kit (Qiagen). The purified PCR products were sequenced in both directions. Using “Sequencher” software (www.msi.umn.edu), the forward and reverse nucleotide sequences were aligned followed by BLAST analysis (www.ncbi.nlm.nih.gov). The nucleotide sequences were then aligned by the Clustal W method using MEGA 4.0 software. The evolutionary distances were computed using the Maximum Composite Likelihood Model. A phylogenetic tree of aligned sequences (based on positions 124 to 467 of AY727882) was then constructed by the neighbour-joining method using 500 bootstrap replicate values. The nucleotide sequences were translated to deduced amino acid sequences to determine the pathotype of NDV involved. The nucleotide sequences were compared with NDV sequences available in GenBank; the virus types and their GenBank accession numbers are presented in . The F gene sequences of different genotypes of class I and class II NDVs were used for comparison. Arabic numerals (1 to 9) are used for indicating class I genotypes, while Roman numerals (I to IX) are used for class II genotypes.
Table 1. Already published NDV F gene sequences used for phylogenetic analysis.
GenBank accession numbers
The NDV sequences of the present study were submitted to the GenBank database with the following accession numbers: NDV-044/09/Bald_Eagle (FJ965310), NDV-046/09/Bald_Eagle (FJ965311), and NDV-045/09/Great horned owl (GQ300974).
Results
Detection of NDV
Using primers specific for the F gene of NDV, a single band of 356 bp was observed with three HA-positive allantoic fluids (termed as NDV isolates) and with the known NDV isolate. Blast analysis of partial sequences of the F gene from all isolates confirmed their identity as NDV. The three NDV isolates were obtained from bald eagles (n=2) and a great horned owl (n=1). Of the three isolates, two were obtained from cloacal samples (great horned owl and bald eagle, one each) and one was from the OP sample of bald eagle. The OP samples corresponding to NDV-positive cloacal samples did not yield NDV. Similarly, the cloacal sample corresponding to the NDV-positive OP sample from bald eagle did not yield NDV. All three NDV-positive birds were sampled in Minnesota.
Cleavage site analysis
All three isolates possessed 112GKQGRL117 at the cleavage site, which is consistent with viruses of low virulence/lentogenic strains.
Phylogenetic analysis
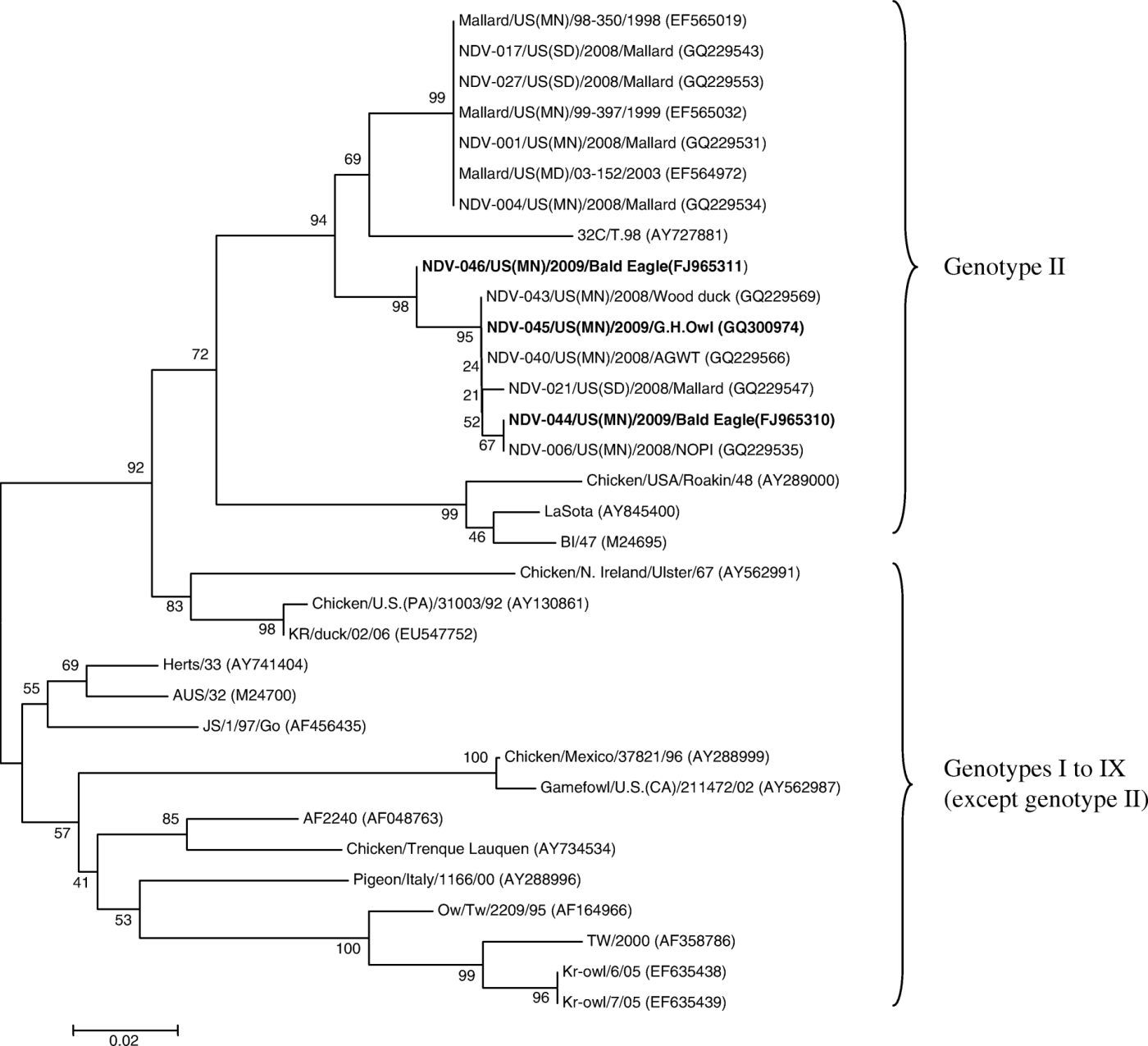
shows the phylogenetic tree based on partial sequences of the F gene together with those from GenBank. All three isolates in this study clustered with published class II genotype viruses. Within class II, the isolates clustered with genotype II NDVs. All virulent strains of class II, used for comparison, were in different clades. Since our isolates clustered with class II genotypes, the class I genotypes initially used for phylogenetic analysis are not shown in . The sequence homology of three NDV isolates was 98.4 to 99.6%. Similarly, the sequence homology with lentogenic strains from wild birds used for comparison varied between 94.5 and 100%. Phylogenetic analysis revealed that NDV isolates from raptors were close to the NDVs from wild birds in the US. The vaccine strains (B1, La Sota) were in a different group from the isolates of the present study.
Figure 1. Phylogenetic tree of the nucleotide sequences of NDV isolates (class II genotypes) based on nucleotides (based on positions 124 to 467 of AY727882) of the coding region of the F gene. GenBank accession numbers are included in parentheses. Strain names from the present study are in bold. Bootstrap values (500 bootstrap replicates) are shown on the tree.
Discussion
The initial aim of the study was to survey raptors for the presence of AIV. No AIV was isolated from the samples tested but NDV was isolated from three out of 120 samples from 60 birds. The isolation of NDV from domestic poultry and waterfowl is common (Marin et al., Citation1996; Takakuwa et al., Citation1998; Huovilainen et al., Citation2001; Jorgensen et al., Citation2004; Seal et al., Citation2005; Kim et al., Citation2007; Jindal et al., Citation2009; Hu et al., Citation2010), but reports on its isolation from raptors are limited (Chu et al., Citation1976; Kou et al., Citation1999; Manvell et al., Citation2000; Lublin et al., Citation2001; Schettler et al., Citation2003; Choi et al., Citation2008). In the present study, we isolated NDV from two species of raptors (i.e. bald eagle and great horned owl). Isolation of NDV from owls has been reported by Kou et al. (Citation1999) in Taiwan. Schettler et al. (Citation2003) have reported on the detection of NDV nucleic acid in different species of raptors such as barn owl, tawny owl, common buzzard, and European kestrel in Germany. Choi et al. (Citation2008) isolated NDV from the Eurasian Scops owl, and Lublin et al. (2001) from the bearded vulture. Wernery et al. (Citation1992) isolated NDV from different species of falcons in the United Arab Emirates. However, there appears to be no report on the isolation of NDV from bald eagles.
All three isolates belonged to class II genotype II NDVs. In an earlier study, we reported the presence of class II NDVs in waterfowl in the Upper Midwest region of the US (Jindal et al., Citation2009). Similarly, Hu et al. (Citation2010) reported only class I NDVs in domestic ducks in China. In contrast, Kim et al. (Citation2007) and Liu et al. (Citation2009) reported the presence of both class I and class II NDVs in wild birds in the US. In the present study we tested a limited number of samples from raptors during a 3-month period. Testing of more samples over a greater period of time may help determine the true prevalence of NDVs circulating in these species. In addition, the possibility of the presence of other genotypes of class II and that of class I in different raptor species cannot be ruled out and merits further study.
The fusion protein is an important determinant for NDV pathogenicity. This protein in velogenic strains of NDV is characterized by the presence of two pairs of basic amino acid residues near the cleavage site. In the lentogenic strains, however, only two single amino acids at the cleavage site are present. Sequences of mesogenic strains contain two pairs of basic amino acid residue or a single arginine and a lysine/arginine pair (Alexander, Citation2003). All three NDV isolates had sequences similar to that of lentogenic strains at their cleavage site. We did not perform intracerebral pathogenicity tests on these isolates; we assume them to be lentogenic in nature based on their cleavage site analysis. Isolation of lentogenic strains of NDV has been reported in waterfowl (Jorgensen et al., Citation2004; Kim et al., Citation2007; Jindal et al., Citation2009) and in raptors (Schettler et al., Citation2003).
It seems probable that NDV pathotypes present in raptors may depend upon the pathotypes present in their prey. The bald eagle, a bird of prey, is found near large bodies of open water. While dependent on fish during the breeding season, bald eagles are opportunistic predators and scavengers throughout the year, feeding especially on waterfowl and deer carcasses throughout the winter (Stalmaster, Citation1987). As waterfowl congregate in large numbers near areas of open water during winter, there are opportunities for transmission of viruses and increasing the exposure probability for eagles that are feeding on them. We believe that feeding on waterfowl carcasses infected with NDV might have led to the transmission of NDV to eagles. Our hypothesis of virus transmission to eagles from waterfowl gains strength from the fact that the NDVs from raptors were phylogenetically closer to published fusion gene sequences of NDVs from waterfowl in the US. Great horned owls have a wide range of prey species including rodents (Stalmaster, Citation1987), which are commonly found around chicken houses as well as waterfowl and upland game birds (e.g. pheasants). In a study, Schettler et al. (Citation2003) identified lentogenic strain (La Sota) from barn owls. The authors were of the view that mice, because of free access to the poultry houses, might be infected or may act as mechanical carriers of vaccine virus; mice are the predominant prey of barn owls. However, the close proximity of the NDV isolate from a great horned owl in this study with NDVs from waterfowl in fusion gene phylogeny suggests the possibility of this species coming in contact with NDV-infected waterfowl.
Velogenic strains of NDV have also been isolated from wild birds (Takakuwa et al., Citation1998; Liu et al., Citation2008; Zhu et al., Citation2010), suggesting an epidemiological link between isolates recovered from outbreaks in poultry with those from wild birds (Takakuwa et al., Citation1998; Huovilainen et al., Citation2001; Jorgensen et al., Citation2004; Kim et al., Citation2007). This signifies the importance of wild birds in the maintenance and transmission of NDVs. We believe that virulent NDVs may be transmitted from waterfowl to raptors, which may spread this virus further due to their wide home range, especially the eagles. Transmission of velogenic strains from raptors to domestic poultry directly or indirectly may lead to outbreaks that can cause considerable economic losses to the poultry industry. Therefore, further epidemiological studies are required to determine the prevalence of different NDV strains in raptors and their impact in relation to transmission to domestic poultry.
Acknowledgements
The present work has been funded in whole or in part with federal funds from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services, under Contract No. HHSN266200700007C. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the NIH.
References
- Alexander , D.J. 2000 . Newcastle disease and other avian paramyxoviruses . Scientific and Technical Review OIE , 19 : 443 – 462 .
- Alexander , D.J. 2003 . “ Newcastle disease, other avian paramyxoviruses, and pneumovirus infection ” . In Diseases of Poultry , 11th edn , Edited by: Saif , Y.M. , Barnes , H.J. , Glisson , J.R. , Fadly , A.M. , McDougald , L.R. and Swayne , D.E. 63 – 92 . Ames : Iowa State University Press .
- Chan , C.H. , Lin , K.L. , Chan , Y. , Wang , Y.L. , Chi , Y.T. Tu , H.L. 2006 . Amplification of the entire genome of influenza A virus H1N1 and H3N2 subtypes by reverse-transcription polymerase chain reaction . Journal of Virological Methods , 136 : 38 – 43 .
- Choi , K.S. , Lee , E.K. , Jeon , W.J. , Nah , J.J. , Kim , Y.J. Lee , M.Y. 2008 . Isolation of a recent Korean epizootic strain of Newcastle disease virus from Eurasian Scops owls affected with severe diarrhea . Journal of Wildlife Diseases , 44 : 193 – 198 .
- Chu , H.P. , Trow , E.W. , Greenwood , A.G. , Jennings , A.R. and Keymer , I.F. 1976 . Isolation of Newcastle disease virus from birds of prey . Avian Pathology , 5 : 227 – 233 .
- Czegledi , A. , Ujvari , D. , Somogyi , E. , Wehmann , E. , Werner , O. and Lomniczi , B. 2006 . Third genome size category of avian paramyxovirus serotype 1 (Newcastle disease virus) and evolutionary implications . Virus Research , 120 : 36 – 48 .
- Dormitorio , T.V. , Giambrone , J.J. , Guo , K. and Hepp , G.R. 2009 . Detection and characterization of avian influenza and other avian paramyxoviruses from wild waterfowl in parts of the southeastern United States . Poultry Science , 88 : 851 – 855 .
- Höfle , U. , Blanco , J.M. and Kaleta , E.F. 2002 . Seroprevalence of avian paramyxovirus 1, 2, and 3 in captive and free-living birds of prey in Spain (preliminary results): implications for management of wild and captive populations . Annals of the New York Academy of Sciences , 969 : 213 – 216 .
- Hu B. Huang Y. He Y. Xu C. Lu X. Zhang W. et al. 2010 Avian influenza virus and Newcastle disease virus (NDV) surveillance in commercial breeding farm in China and the characterization of class I NDV isolates Veterinary Microbiology PMID 20133090 , 144 82 86
- Huovilainen , A. , Ek-Kommonen , C. , Manvell , R. and Kinnunen , L. 2001 . Phylogenetic analysis of avian paramyxovirus 1 strains isolated in Finland . Archives of Virology , 146 : 1775 – 1785 .
- Jindal N. Chander Y. Chockalingam A.K. de Abin M. Redig P.T. Goyal S.M. 2009 Phylogenetic analysis of Newcastle disease viruses isolated from waterfowl in the Upper Midwest Region of the United States Virology Journal 6 191 (http://www.virologyj.com/content/6/1/191 )
- Jorgensen , P.H. , Handberg , K.J. , Ahrens , P. , Therkildsen , O.R. , Manvell , R.J. and Alexander , D.J. 2004 . Strains of avian paramyxovirus type 1 of low pathogenicity for chickens isolated from poultry and wild birds in Denmark . The Veterinary Record , 154 : 497 – 500 .
- Keymer , I.F. and Dawson , P.S. 1971 . Newcastle disease in birds of prey . The Veterinary Record , 88 : 432
- Kim , L.M. , King , D.J. , Curry , P.E. , Suarez , D.L. , Swayne , D.E. Stallknecht , D.E. 2007 . Phylogenetic diversity among low-virulence Newcastle disease viruses from waterfowls and shorebirds and comparison of genotype distributions to those of poultry-origin isolates . Journal of Virology , 81 : 12641 – 12653 .
- Kocan , A.A. , Snelling , J. and Greiner , E.C. 1977 . Some infectious and parasitic diseases in Oklahoma raptors . Journal of Wildlife Diseases , 13 : 304 – 306 .
- Kou , Y.T. , Chueh , L.L. and Wang , C.H. 1999 . Restriction fragment length polymorphism analysis of the F gene of Newcastle disease viruses isolated from chickens and an owl in Taiwan . Journal of Veterinary and Medical Science , 61 : 1191 – 1195 .
- Liu , H. , Wang , Z. , Wang , Y. , Sun , C. , Zheng , D. and Wu , Y. 2008 . Characterization of Newcastle disease virus isolated from waterfowl in China . Avian Diseases , 52 : 150 – 155 .
- Liu , X. , Wang , X. , Wu , S. , Hu , S. , Peng , Y. , Xue , F. and Liu , X. 2009 . Surveillance of avirulent Newcastle disease viruses in domestic ducks (Anas platyrhynchos and Cairina moschata) at live bird markets in Eastern China and characterization of the viruses isolated . Avian Pathology , 38 : 377 – 391 .
- Lublin , A. , Mechani , S. , Siman-Tov , Y. , Weisman , Y. , Horowitz , H.I. and Hatzofe , O. 2001 . Sudden death of a bearded vulture (Gypaetus barbatus) possibly caused by Newcastle disease virus . Avian Diseases , 45 : 741 – 744 .
- Manvell R.J. Wernery U. Alexander D.J. Frost K.M. 2000 Newcastle disease (Avian PMV-1) in raptors In Proceedings of the 3rd International Raptor Biomedical Conference. Raptor Biomedicine III 3 8 Zoological Education Network, Fort Worth, Texas, USA
- Marin , M.C. , Villegas , P. , Bennett , J.D. and Seal , B.S. 1996 . Virus characterization and sequence of the fusion protein gene cleavage site of recent Newcastle disease virus field isolates from the southeastern United States and Puerto Rico . Avian Diseases , 40 : 382 – 390 .
- Nanthakumar , T. , Kataria , R.S. , Tiwari , A.K. , Butchaiah , G. and Kataria , J.M. 2000 . Pathotyping of Newcastle disease viruses by RT-PCR and restriction enzyme analysis . Veterinary Research Communications , 24 : 275 – 286 .
- Pedersen , J.C. , Senne , D.A. , Woolcock , P.R. , Kinde , H. , King , D.J. Wise , M.G. 2004 . Phylogenetic relationships among virulent Newcastle disease virus isolates from the 2002–2003 outbreak in California and other recent outbreaks in North America . Journal of Clinical Microbiology , 42 : 2329 – 2334 .
- Schettler , E. , Fickel , J. , Hotzel , H. , Sachse , K. , Streich , W.J. , Wittstatt , U. and Frolich , K. 2003 . Newcastle disease virus and Chlamydia psittaci in free-living raptors from Eastern Germany . Journal of Wildlife Diseases , 39 : 57 – 63 .
- Schettler , E. , Langgemach , T. , Sommer , P. , Streich , J. and Frolich , K. 2001 . Seroepizootiology of selected infectious disease agents in free-living birds of prey in Germany . Journal of Wildlife Diseases , 37 : 145 – 152 .
- Seal , B.S. , Wise , M.G. , Pedersen , J.C. , Senne , D.A. , Alvarez , R. Scott , M.S. 2005 . Genomic sequences of low-virulence avian paramyxovirus-1 (Newcastle disease virus) isolates obtained from live-bird markets in North America not related to commonly utilized commercial vaccine strains . Veterinary Microbiology , 106 : 7 – 16 .
- Stalmaster , M.V. 1987 . The Bald Eagle , New York : Universe Publishing Co .
- Takakuwa , H. , Ito , T. , Takada , A. , Okazaki , K. and Kida , H. 1998 . Potentially virulent Newcastle disease viruses are maintained in migratory waterfowl populations . Japanese Journal of Veterinary Research , 45 : 207 – 215 .
- Wernery , U. , Remple , J.D. , Neumann , U. , Alexander , D.J. , Manvell , R.J. and Kaaden , O.R. 1992 . Avian paramyxoviruses serotype 1 (Newcastle disease virus) – infections in falcons . Journal of Veterinary Medicine B , 39 : 153 – 158 .
- Wise , M.G. , Suarez , D.L. , Seal , B.S. , Pedersen , J.C. , Senne , D.A. King , D.J. 2004 . Development of a real-time reverse-transcription PCR for detection of Newcastle disease virus RNA in clinical samples . Journal of Clinical Microbiology , 42 : 329 – 338 .
- Zhu , W. , Dong , J. , Xie , Z. , Liu , Q. and Khan , M.I. 2010 . Phylogenetic and pathogenic analysis of Newcastle disease virus isolated from house sparrow (Passer domesticus) living around poultry farm in southern China . Virus Genes , 40 : 231 – 235 .