Abstract
As part of an epidemiological study of infectious bronchitis virus (IBV) in Brazil, 252 samples from IBV-suspect flocks were tested and the IBV-positive samples were analysed by sequencing of hypervariable regions 1 and 2 of the S1 gene. A high prevalence of IBV variants was found and the sequence analysis of 41 samples revealed a high molecular similarity among the Brazilian isolates (from 90.2 to 100% and from 85.3 to 100% nucleotide and amino acid identity, respectively). The Brazilian isolates showed low genetic relationship with Massachusetts (63.4 to 70.7%), European (45.9 to 75.6%), American (49.3 to 76.4%) and other reference serotypes (67.5 to 78.8%). The Brazilian isolates branched into one unique cluster, separate from the reference serotypes used for infectious bronchitis control in other countries. The variants analysed in this work had a high similarity with all previously published Brazilian IBV isolates, suggesting the presence and high prevalence of a unique or predominant genotype circulating in Brazil. In addition, the virus neutralization test showed that the three Brazilian isolates analysed in the present study are antigenically related to one another but are different from the Massachusetts serotype. The present study shows that IBVs of a unique genotype can be associated with different clinical diseases, and that low genetic variation was detected in this genotype over a long period of time. The molecular characterization of the Brazilian variants isolated from 2003 to 2009 from different geographic regions of the country shows that only one predominant genotype is widespread in the Brazilian territory, denominated in this study as BR-I genotype.
Introduction
Infectious bronchitis (IB) is one of the most economically significant diseases of the poultry industry and can be involved in respiratory disease, nephritis, enteritis, fertility problems and reduced egg production and quality. The disease is caused by infectious bronchitis virus (IBV), a member of the Coronaviridae family (Ignjatovic et al., Citation2006; Cavanagh & Gelb, Citation2008).
IBV is primarily a respiratory pathogen because it replicates mainly in epithelial cells of the trachea, but it can also replicate in other respiratory and non-respiratory tissues such as the kidney, gonad and alimentary tract. Thus, nephropathogenic and field strains causing reduction of fertility in cockerels, damage to the female reproductive tract and enteric disease have been described (Ambali & Jones, Citation1990; Boltz et al., Citation2004; Liu & Kong, Citation2004; Villarreal et al., Citation2007a; Cavanagh & Gelb, Citation2008).
The IBV genome is a single-stranded linear RNA molecule, and its virion contains four structural proteins. The S glycoprotein is proteolytically processed into two non-covalently bound peptide chains known as S1 and S2. The S1 subunit contains epitopes and determinants for virus neutralizing antibodies, protective immunity, cell attachment and serotype specificity (Ignjatovic & Galli, Citation1994). Therefore, analysis of the S1 gene has been used to differentiate IBV genotypes and serotypes. Genotyping, which may correlate with the serotype, is frequently used because it is extremely fast and convenient and is a better predictor of protection than serotyping by haemagglutination inhibition or virus neutralization (VN) tests (Wang & Huang, Citation2000; Bochkov et al., Citation2006; Ladman et al., Citation2006). Reverse transcriptase-polymerase chain reaction (RT-PCR) followed by restriction fragment length polymorphism (RFLP) analysis of the amplified S1 gene and detection of genotype-specific regions of the genome by multiplex RT-PCR have been widely used to differentiate vaccine and field strains (Kwon et al., Citation1993; Cavanagh et al., Citation1999; Lee et al., Citation2003; Liu et al., Citation2003; Zanella et al., Citation2003). Nevertheless, nucleotide sequencing of the S1 gene region, including hypervariable region (HVR) 1 and HVR 2, is widely recognized as the most useful technique for the differentiation of IBV strains (Ignjatovic et al., Citation2006; Worthington et al., Citation2008).
Vaccination is only partially successful because of the continual emergence of antigenic variants (Ignjatovic & Sapats, Citation2000). This IBV evolution can be explained by the continuing use of live vaccines, immunological pressure exerted on circulating viruses by the continuing presence of an immune bird population and changes in the viral RNA due to nucleotide insertions, deletions, or point mutations in the S1 gene resulting from errors made by the viral polymerase (Ignjatovic et al., Citation2006). Consequently, vaccination programmes must include previous identification of the IBV strain prevalent in the field (Cavanagh et al., Citation2005; Cavanagh & Gelb, Citation2008). Molecular epidemiological studies are then necessary to determine the genotypes circulating in the region or country and to select the appropriate vaccine for IB control in each geographical region or country.
Vaccination against IBV in Brazil is carried out only with live and inactivated vaccines containing virus of the Massachusetts (Mass) serotype, although field observations suggest poor or no protection against wild isolates because IBV outbreaks are frequently observed in vaccinated and non-vaccinated farms. Antigenic groups different from the Mass serotype have already been described in this country (Di Fabio et al., Citation2000). Recently, IBV variant strains isolated from birds showing enteric disorders in broilers and fertility disorders in cockerals have been described in Brazil (Villarreal et al., Citation2007a, Citation2007b; Chacón et al., Citation2009). However, epidemiological studies that include a large number of samples collected from chickens with different clinical manifestations and from different geographic regions have not been carried out. To establish which IBV genotypes are circulating in Brazil, IBVs collected during a period of 7 years from different clinical diseases and from the nine main poultry-producing regions were characterized by PCR-RFLP and sequencing of the S1 protein gene. In addition, based on the S1 gene sequence analysis, three field isolates were selected to be used in the VN test.
Materials and Methods
Reference viruses
Mass vaccine strains (Fort Dodge Animal Health, Merial and Ceva Animal Health Laboratories, Brazil) and RNA from Connecticut, Arkansas, D274 and 793/B (otherwise called 4/91) serotypes were used as controls in the molecular assays.
Field samples
Samples from 252 commercial farms in Brazil showing clinical signs suggestive of IB were collected between 2003 and 2009. The samples were collected from different geographic regions: North East (Bahia and Pernambuco), Centre-southeast (Espírito Santo, Minas Gerais, Mato Grosso and São Paulo), and South (Paraná, Rio Grande do Sul and Santa Catarina) and sent to the Avian Pathology Laboratory at the University of São Paulo for virological examination. Both vaccinated and non-vaccinated broilers, hens, breeders and grandparents were included in the present study. Birds of different ages and with respiratory signs, low growth rates, enteric disorders, low egg production and low fertility (females and males) were included. In several farms, mortality was observed at rates ranging from 2 to 15%. Trachea, lungs and kidneys were collected from all of the analysed flocks, while the enteric contents and reproductive organs were collected from flocks with digestive and reproductive disorders, respectively.
Sample preparation and RNA extraction
Tissue samples from five birds of each farm were homogenized in sterile phosphate-buffered saline, pH 7.4, 0.01 M, making 20% (w/v) suspensions that were clarified by centrifugation at 3000×g for 30 min at 4°C. RNA was extracted from the supernotants with TRIzol reagents (Invitrogen™) according to the manufacturer's recommendations.
IBV detection
For IBV screening, viral RNA extracted from tissues from each of the 252 commercial farms was analysed using primers designed to amplify a fragment of the untranslated region of IBV. The reaction conditions and primers described by Cavanagh et al. (Citation2002) were used for this purpose.
IBV typing by multiplex RT-nested PCR
To differentiate between the Mass serotype and the Brazilian variants, the positive samples in the screening assay were analysed by a multiplex RT-nested PCR, which can differentiate Mass from other strains. The primers XCE1+, XCE2−, XCE3−, MCE1+ (specific for Mass), BCE1+ (793B) and DCE1+ (D274) were used as described previously (Cavanagh et al., Citation1999; Zanella et al., Citation2003). The samples that did not produce a fragment of 295 bp corresponding to the Mass serotype were considered variant strains.
Virus growth and titration
Fifty IBV variant samples were propagated in five 9-day-old specific pathogen free chicken embryos (Laboratório Biovet, Vargem Grande Paulista, Brazil) before DNA sequencing. Two to three passages were performed by inoculating the harvested allantoic fluids into five other eggs until characteristic IB lesions were observed. The allantoic fluids of the eggs from the last passage were harvested for molecular analysis. In addition, the sixth passage of samples USP-11 and USP-50 were titrated by the Reed and Muench method (Villegas, Citation1998) and used to perform the VN test (see below).
RT-PCR-RFLP
Extracted RNA from eight variants and IB reference strains was analysed by RT-PCR-RFLP analysis. This technique was carried out using the primers S1OLIGO5′ and S1OLIGO3′, as described by Kwon et al. (Citation1993). The amplified products of ~1700 nucleotides, which included the entire S1 gene, were digested with the endonucleases BstYI, HaeIII and XcmI (New England BioLabs, Beverly, Massachusetts, USA). Then, RFLPs were visualized by agarose gel electrophoresis with ethidium bromide.
Amplification and sequencing of the S1 gene
Forty-one Brazilian isolates, differentiated from the Mass serotype by multiplex RT-nested PCR, were further analysed after amplification and sequencing of a fragment of the S1 gene containing HVR 1 and HVR 2. The RT-PCR was performed as described by Gelb et al. (Citation2005) using the primers S1 OLIGO5′ and CK2 (Kwon & Jackwood, Citation1995; Keeler et al., Citation1998). The PCR products were analysed on a 1.5% agarose gel and purified using a GFX™ PCR DNA and Gel Band Purification Kit (GE Healthcare, Piscataway, New Jersey, USA) as described by the manufacturer. Each purified product was sequenced in the forward and reverse directions according to the instructions of the BigDye Terminator v3.1 Cycle Sequencing kit (Applied Biosystems, Foster City, California, USA). Sequencing reactions were run in an ABI PRISM 3730 Genetic Analyzer (Applied Biosystems).
Nucleotide and amino acid sequence analysis
The analysed fragment included positions 1 to 535 of the H120 strain S1 gene sequence (Genbank accession number M21970). Comparative analysis of nucleotide and deduced amino acid sequences from Brazilian and reference strains was performed with the CLUSTAL W method available in the Bioedit software package. The phylogenetic tree was obtained using the neighbour-joining method with 1000 bootstrapping replicates integrated in the MEGA software version 3.1 (Kumar et al., Citation2004).
Genbank accession numbers
The S1 sequence data of Brazilian IBV isolates previously published in Genbank and used in the phylogenetic analysis included: USP-1 (DQ355995), USP-02 (DQ448273), USP-3 (DQ448277), USP-4 (DQ492307), USP-5 (DQ492308), USP-6 (DQ492309), USP-7 (DQ448274), USP-8 (DQ492310), USP-9 (DQ492311), USP-10 (DQ448275), USP-11 (DQ492312), and USP-12 (DQ448276). In addition, sequences of reference strains used for comparison in the present study were obtained from the following Genbank database accession numbers: CO1692 (AY604547), CO8250 (AY604552), CO8232 (AY604558), CO8089 (AY604553), AR/03/BA/06 (FJ167386), AR/06/BA/13 (FJ167376), AR/06/BA/14 (FJ167375), Beaudette (X02342), H120 (M21883), H52 (AF352315), M41 (X04722), Connecticut (L18990), Arkansas99 (L10384), Ark/15c/96 (AF169859), Delaware (U77298), GA/2787/98 (AF274438), JMK (L14070), Gray (L14069), Florida 18288 (AF27512), Holte (L18988), Qu16 (AF349620), Qu mv (AF349621), 4/91 attenuated (AF093793), 4/91 pathogenic (AF093794), UK/7/91-793/B (Z83975), D274 (X15832), D1466 (M21971), Italy02 (AJ457137), Spain/99/316 (DQ064809), FR/L-1450L/05 (EF079117), RF/01/99 (AJ440783), Israel/720/99 (AY91552), K507-01 (AY257064), K774-01 (AY257065), QX (AF193423), JK/99/01 (AF210735), A2 (AY43312), T Australia (AY775779), VicS (U29519), N1/62 (U29522), N4/02 (DQ059618), New Zealand (AF151958), and Egypt/F/03 (DQ487085).
VN test
Three variant viruses were selected for the VN test. Two (USP-11 and USP-50) were tested against sera produced to another Brazilian field virus (USP-40). Monospecific antiserum to isolate USP-40 was prepared as previously described (Gelb & Jackwood, Citation1998). Briefly, specific pathogen free chickens were inoculated with approximately log10 5 median embryo infectious doses per bird by the intratracheal and intravenous route at 3 and 5 weeks of age. Blood was obtained 3 weeks after the last inoculation, and serum was harvested and inactivated at 56oC for 30 min. Then, antisera to isolate USP-40 and serotype Mass (Charles River Laboratories, North Franklin, Connecticut, USA) were used in the VN test that was performed using the diluted-serum constant virus method (beta procedure) as described by Thayer & Beard (Citation1998). Briefly, four-fold serial dilutions of each antiserum were reacted in equal volumes with virus suspension containing log10 2.0 median embryo infectious doses/0.1 ml and incubated at room temperature for 1 h. Each serum–virus mixture was inoculated into specific pathogen free chicken embryonated eggs via the allantoic route. Chicken embryos that died 24 h after inoculation were considered non-specific mortality. Embryos were evaluated 1 week after inoculation for the presence of IBV-specific lesions representing non-virus neutralization. Endpoint titres were calculated by the Reed and Muench method.
Results
IBV detection
From the 252 samples analysed, IBV was detected in 212 commercial farms (84%) using the RT-nested PCR assay. IBV was detected in broilers, layers, breeders and grandparents from both vaccinated and non-vaccinated farms. IBV was identified in samples from all geographic regions collected between 2003 and 2009.
IBV typing
Of the 212 IBV samples, 175 (83%) were classified as variants by the multiplex RT-nested PCR because these samples did not show the band specific to the Mass serotype (295 bp). Similarly, the multiplex RT-PCR assay indicated that the Brazilian samples were different from the 793/B (4/91) and D274 vaccine strains. Variants were detected in broiler, layer, breeder and grandparent farms from all nine geographic regions included in the study. In 158 cases (90%), variant isolates were detected in vaccinated farms. The birds had been vaccinated with at least one live vaccine dose, sometimes in combination with inactivated vaccines. Interestingly, IBV variants were detected from farms with chickens showing different clinical signs ().
Table 1. Geographic origin and clinical signs associated with IBV variants detected and characterized in the present study
PCR-RFLP analysis
This analysis was conducted to differentiate Brazilian variants and reference strains. Eight Brazilian variants were submitted to RFLP analysis. All of the Brazilian isolates had RFLP patterns that were different to the Mass, Connecticut, Arkansas, 793/B and D274 strains when digested by BstYI, HaeIII and XcmI. The Brazilian isolates showed three different patterns using BstYI and HaeIII: pattern A (USP-34, USP-36 and USP-40), pattern B (USP-41, USP-49 and USP-50), and pattern C (USP-51 and USP-52).
S1 amplification and sequencing
A fragment of approximately 700 bp of the S1 gene of 41 Brazilian isolates obtained from broiler, layer, breeder and grandparent birds with different clinical manifestations were amplified and sequenced. The 41 sequenced samples were recovered from the four main chicken-producing Brazilian States (). The nucleotide sequence data reported in this paper are available in Genbank under the following accession numbers: USP-33 (GU383070), USP-34 (GU383071), USP-35 (GU383072), USP-36 (GU383073), USP-37 (GU383074), USP-38 (GU383075), USP-39 (GU383076), USP-40 (GU383077), USP-41 (GU383078), USP-42 (GU383079), USP-43 (GU383080), USP-44 (GU383081), USP-45 (GU383082), USP-46 (GU383083), USP-47 (GU383084), USP-48 (GU383085), USP-49 (GU383086), USP-50 (GU383087), USP-51 (GU383088), USP-52 (GU383089), USP-53 (GU383090), USP-54 (GU383091), USP-55 (GU383092), USP-56 (GU383093), USP-57 (GU383094), USP-58 (GU383095), USP-59 (GU383096), USP-60 (GU383097), USP-61 (GU383098), USP-62 (GU383099), USP-63 (GU383100), USP-64 (GU383101), USP-65 (GU383102), USP-66 (GU383103), USP-67 (GU383104), USP-68 (GU383105), USP-69 (GU383106), USP-70 (GU383107), USP-71 (GU383108), USP-72 (GU383109), and USP-73 (GU383110).
Table 2. Epidemiological information from Brazilian IBV field isolates sequenced in the present study (BR-I genotype).
Phylogenetic analysis
Nucleotide and amino acid sequences of the S1 gene from 41 Brazilian IBV isolates obtained in the present study were aligned and compared with previously published Brazilian and known reference strains worldwide. The analysis revealed that all sequences obtained in this study were closely related genetically. The nucleotide and amino acid sequence analysis of the 41 Brazilian isolates yielded identity values between 90.3 and 100% and between 90.2 and 100%, respectively. The phylogenetic analysis revealed 68.3 to 71.9% amino acid similarity between the Brazilian isolates and the Mass serotype. Brazilian isolates also showed a low similarity with reference serotypes: 45.9 to 75.6% identity between Brazilian isolates and European serotypes (including D1466, D274, 793/B and Italy 02); 49.3 to 76.4% identity with American serotypes (including Connecticut, Arkansas, Delaware, GA, JMK, Gray, Florida and Holte); 66.8 to 69.8% identity with Asiatic genotypes (including QX, JK/99/01, K507-01 and K774-01); 59.4 to 78.8% identity with Australian/Pacific area strains (including T Australian, N4/02, VicS and K63 New Zealand); and 68.9 to 72.5% identity with the African genotype (Egypt/F/03) (). The sequences obtained in the present study were closely related to other Brazilian IBV isolates previously published (90.2 to 100% identity). On the other hand, the Brazilian isolates had 82 to 88% and 44 to 75% genetic relationship with Argentinean and Colombian IBVs, respectively. All Brazilian samples were grouped within the same genotypic cluster, well separated from genotypes from other countries and from the reference strains used for vaccination worldwide (). Subgroups could not be well defined in the Brazilian cluster based on the geographic origin, year of isolation or type of clinical disease observed in the field.
Figure 1. Phylogenetic relationships of the Brazilian IBV isolates and reference strains based on S1 gene amino acid sequences determined using MEGA 3.1 with the Clustal W method. Numbers along the branches refer to bootstrap values; the bar represents the number of substitutions per site. *Sequence previously published in GenBank.
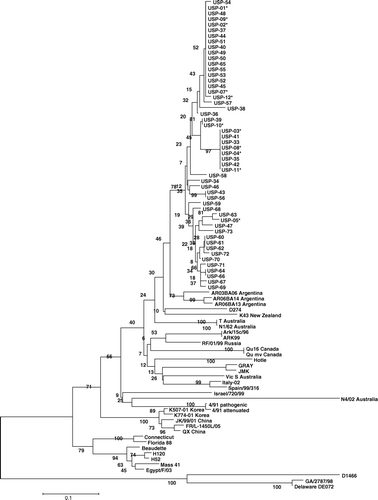
Table 3. Comparison of the nucleotide and amino acid sequences from the S1 gene of Brazilian isolates (2003 to 2009) and reference IBV strains.
VN analysis
Based on S1 partial sequencing, three field isolates were selected for use in the VN test. Viruses were considered of the same serotype if the serum protected 50% or more of the embryos. The VN test results showed little antigenic relationship (29 to 33%) between the Brazilian isolates and the Mass serotype (28.6 to 33.2%), but a high antigenic relationship among the Brazilian viruses (94.7 to 98.2%).
Discussion
Despite the vaccination schemes that include at least one live vaccine in broilers and at least two live and one inactivated vaccine in layers and breeders using the Mass strain, IB outbreaks have been reported frequently in vaccinated and non-vaccinated chicken flocks in Brazil. In addition, IBV is detected in birds with different clinical manifestations including respiratory disease, reduction in fertility, lower egg production, enteric disorders and poor weight gain. These field observations suggested that one or more genotypes/serotypes that are genetically and antigenically divergent from the Mass vaccine strain might be circulating in Brazilian farms.
IBV variants have been reported in this country (Villarreal et al., Citation2007b; Chacón et al., Citation2009) and the viruses characterized in these studies shown to be genetically similar. However, they originated from only two geographic regions and were isolated mainly from broiler chickens showing enteric disorders. In this study we characterized viruses isolated from different types of bird showing different clinical diseases and from a higher number of geographic regions.
In the present work, 252 field samples collected from nine geographic states over a 7-year period were investigated by IBV molecular detection and characterization. IBV was detected in 212 farms (84%), and the assay used for typing revealed that 83% of the detected IBVs (175 viruses) were not related to the Mass strain. The 175 variant IBVs were detected in all geographic regions analysed, and were recovered from broilers, layers, breeders and grandparents showing different clinical manifestations. The high prevalence of variants in vaccinated farms shows the failure of the Mass serotype vaccines to protect the chickens against challenge with the Brazilian variants.
The isolation and typing of the IBV field isolates is necessary, not only for the study of virus evolution, but also for effective modification of vaccination programs (Cavanagh et al., Citation2005). For this reason, 41 variant viruses taken from different geographic regions, types of birds, clinical manifestations and years of isolation were randomly selected for partial sequencing of the S1 gene and for PCR-RFLP analysis (Kwon et al., Citation1993 ; Gelb et al., Citation2005). The phylogenetic analysis showed a high genetic relationship among all variant strains analysed, which are hereafter referred to as the Brazil-I (BR-I) genotype (). In addition, the variant isolates were closely related to all of the Brazilian strains previously published in Genbank. The circulation of such a high number of variant isolates of a unique genotype indicates that they probably originated from the same source. Interestingly, it was not possible to differentiate among samples recovered from different geographic regions, indicating the widespread nature of one predominant variant throughout the Brazilian territory, because genetically similar viruses were found in regions as far from one another as 2200 km. Di Fabio et al. (Citation2000) reported the presence of at least four antigenic types divergent from Mass serotype in Brazil in samples collected in 1995. However, in that study the authors reported that chickens vaccinated with Mass serotype were well protected against the challenge using isolates of three of these antigenic groups. Furthermore, changes in the prevalence of genotypes in a geographic area (i.e. their emergence, disappearance and dissemination) have already been observed (Dolz et al., Citation2006; Worthington et al., Citation2008). Although the presence of more than one variant serotype cannot be excluded, the results obtained in this study show that only one predominant genotype was circulating in Brazil in recent years.
The presence and spread of some IBV strains to close geographical regions have been described (Ignjatovic & Sapats, Citation2000; Meulemans et al., Citation2001; Ignjatovic et al., Citation2006). Moreover, the detection of genotypes over a long distance (e.g. in different continents) has not yet been described. Only the QX strain has been isolated in very distant territories (Meulemans et al., Citation2001; Bochkov et al., Citation2006; Worthington et al., Citation2008). Isolates genetically related to the BR-I genotype were detected in Argentina (Rimondi et al., Citation2009). The fact that the BR-I genotype is widespread in Brazil suggests that this virus could have been disseminated to Argentina. This finding shows that the BR-I genotype is widely distributed and may be present in other neighbouring countries. Nevertheless, the Brazilian isolates exhibited a low genetic similarity with Colombian variants. Colombia and Argentina are neighbouring countries to Brazil, but the Brazilian poultry industry is located closer to the Argentinean territory. In addition, geographical barriers limit commercial relationships between countries; consequently they also limit the dissemination of avian pathogens. Indeed, commercial relationships that are more intensive between Brazil and Argentina favoured the spreading of the BR-I genotype. The risk factor associated with intensive commercial relations had already been reported by other authors (Bochkov et al., Citation2006).
When eight Brazilian variants were submitted to PCR-RFLP analysis, three patterns were distinguished. Nevertheless, the eight isolates showed high nucleotide and amino acid similarity in the S1 gene. Probably, point mutations exist at BstYI and HaeIII restriction sites in the analysed samples. Discrepancies between RFLP and sequencing results were observed previously by Gelb et al. (Citation2005). In that study, the sequencing findings were supported by results of assessing immunity after challenge. We observed that characterization using the partial sequence of the S1 gene offers better results than PCR-RFLP because viruses with small differences in the nucleotide sequence show different RFLP patterns.
The S1 gene contains determinants for cell tropism (Cavanagh et al., Citation2005), and thus differences in this gene might be found in strains with different cell tropisms. Brazilian isolates recovered from birds with different clinical signs showed a high molecular similarity ( and ). This finding is in agreement with Zanella et al. (Citation2003), who observed that one virus can produce different clinical signs in different types of chickens. In addition, the QX strain was detected in China and associated with proventriculitis, but in Europe the QX strain was isolated from cases of nephritis and from false layers (Worthington et al., Citation2008). The results of the present study show for the first time that viruses of the same genotype can be associated with multiple clinical conditions. Probably, the differences observed in the field depend on other factors such as the age and physiological states of the chickens at the time of infection or complications due to secondary pathogens (Boshkov et al., 2006). Our field observations suggest that there is a higher virus replication and consequently more severe lesions in tissues where there are epithelial cells in intensive multiplication due to physiological demand.
It has been reported that the pressure caused by the introduction of new vaccine serotypes affects the speed of evolution of IBV in the field (Ignjatovic et al., Citation2006). The close genetic relationship among all Brazilian isolates collected during a 7-year period suggests that the BR-I genotype is apparently stable over time in the field. It seems that the Mass serotype, which was the only vaccine serotype introduced in Brazil after the first detection of IBV in this country, does not exert pressure on the evolution of the Brazilian BR-I genotype. In addition, a small amount of variation in the S1 gene (up to approximately 6%) has been observed in other serotypes over long periods of time (Cavanagh et al., Citation2005).
The best protection against a challenge by IBV is provided by a vaccine containing the homologous strain, and vaccines containing heterologous strains may not provide adequate cross-protection (Gelb et al., Citation1991). The phylogenetic analysis revealed a low genetic similarity between the Brazilian variant and the Mass serotype. In addition, the VN test showed low antigenic relationships between the Brazilian isolates and the Mass strain, and showed that the three Brazilian isolates are antigenically related. These findings explain the failure of the vaccination strategies used in Brazil, since the BR-I variant was isolated from birds vaccinated with the Mass serotype.
Another important finding of the present work was that the Brazilian IB variants are markedly different from all other known IB viruses, and thus the introduction of IBV vaccines foreign to this region is not advisable. Di Fabio et al. (Citation2000) have already shown that Brazilian IBVs were different serologically from those reported in other countries. The combination of live vaccines containing two different serotypes is suggested in territories with a broad genetic variability (Bochkov et al., Citation2006). However, this is not the case in Brazil.
Sequence analysis of the S1 gene showed that Brazilian isolates form a separate phylogenetic group, showing distant genetic relationships with reference vaccine strains. All Brazilian variants showed a high similarity, indicating that they all belong to the same genotype. However, molecular studies have shown that a new serotype can emerge as the result of only a few changes in the amino acid composition of the S1 subunit (Cavanagh et al., Citation1992). Therefore, additional serological tests should be used to confirm that all or the majority of genotype I isolates belong to one unique serotype.
In summary, the present study shows that one predominant Brazilian genotype, the “BR-I genotype”, is widespread in Brazil, as it has been found in very distant territories including a neighbouring country. Phylogenetic analysis revealed genetic stability of this genotype over time, since a high similarity was found in viruses isolated during a period of 7 years. The detection of this genotype in broiler, layer, breeder and grandparent commercial flocks showing different clinical manifestations demonstrates that a genotype might be associated with multiple clinical conditions. The S1 gene analysis revealed a low genetic similarity between the BR-I genotype and Mass or other reference strains, while the VN test showed that viruses of BR-I genotype are different antigenically from the Mass serotype.
Acknowledgements
The authors would like to thank Laboratório Biovet, Fort Dodge Animal Health, Ceva Animal Health and Merial (Brazil) for providing the embryonated eggs and vaccine strains; the veterinarians and farmers who submitted clinical samples; and the Fundação de Amparo à Pesquisa do Estado de São Paulo) and the Conselho Nacional de Desenvolvimento Científico e Tecnológico for financial support (grant 06/59332-9).
References
- Ambali , A.G. and Jones , R.C. 1990 . Early pathogenesis in chicks of infection with an enterotropic strain of infectious bronchitis virus . Avian Diseases , 34 : 809 – 817 .
- Bochkov , Y.A. , Batchenko , G.V. , Shcherbakova , L.O. , Borisov , A.V. and Drygin , V.V. 2006 . Molecular epizootiology of avian infectious bronchitis in Russia . Avian Pathology , 35 : 379 – 393 .
- Boltz , D.A. , Nakai , M. and Bahr , J.M. 2004 . Avian infectious bronchitis virus: a possible cause of reduced fertility in the rooster . Avian Diseases , 48 : 909 – 915 .
- Cavanagh , D. and Gelb , J. 2008 . “ Infectious bronchitis ” . In Diseases of Poultry , 12th edn , Edited by: Saif , Y.M. , Barnes , H.J. , Glisson , J.R. , Fadly , A.M. , McDougald , L.R. and Swayne , D.E. 117 – 135 . Ames : Iowa State Press .
- Cavanagh , D. , Davis , P.J. and Cook , J.K.A. 1992 . Infectious bronchitis virus: evidence for recombination within the Massachusetts serotype . Avian Pathology , 21 : 401 – 408 .
- Cavanagh , D. , Mawditt , K. , Britton , P. and Naylor , C.J. 1999 . Longitudinal field studies of infectious bronchitis virus and avian pneumovirus in broilers using type-specific polymerase chain reactions . Avian Pathology , 25 : 593 – 605 .
- Cavanagh , D. , Mawditt , K. , Welchman , D.B. , Britton , P. and Gough , R.E. 2002 . Coronaviruses from pheasants (Phasianus colchicus) are genetically closely related to coronaviruses of domestic fowl (infectious bronchitis virus) and turkeys . Avian Pathology , 31 : 81 – 93 .
- Cavanagh , D. , Picault , J.P , Gough , R. , Hess , M. , Mawditt , K. and Britton , P. 2005 . Variation in the spike protein of the 793/B type of infectious bronchitis virus, in the field and during alternate passage in chickens and embryonated eggs . Avian Pathology , 34 : 20 – 25 .
- Chacón , J.L.V , Assayag , M.S. , Revolledo , L. , Ivo , M. , Vejarano , M.P. , Pedroso , A.C. , & Ferreira , A.J.P. 2009 . Pathogenic patterns in chicken challenged with variant strains of infectious bronchitis virus isolated from chicken flocks with different clinical manifestations . In Proceedings of the VIth International Symposium on Avian Corona- and Pneumoviruses 138 141 . Rauischholzhausen, Germany .
- Di Fabio , J. , Rossini , L.I. , Orbell , S.J. , Paul , G. , Huggins , M.B. Malo , A. 2000 . Characterization of infectious bronchitis virus isolated from outbreaks of disease in commercial flocks in Brazil . Avian Diseases , 44 : 582 – 589 .
- Dolz , R. , Pujols , J. , Ordoñez , G. , Porta , R. and Majo , N. 2006 . Antigenic and molecular characterization of isolates of the Italy 02 infectious bronchitis virus genotype . Avian Pathology , 35 : 77 – 85 .
- Gelb , J. and Jackwood , M.W. 1998 . “ Infectious bronchitis ” . In A Laboratory Manual for the Isolation and Identification of Avian Pathogens , 4th edn , Edited by: Swayne , D.E. , Glisson , J.R. , Jackwood , M.W. , Pearson , J.E. and Reed , W.M. 169 – 173 . Pennsylvania, PA : The American Association of Avian Pathologists .
- Gelb , J. , Wolff , J.B. and Moran , C.A. 1991 . Variant serotypes of infectious bronchitis virus isolated from commercial layer and broiler chickens . Avian Diseases , 35 : 82 – 87 .
- Gelb , J. , Weisman , Y. , Ladman , B.S. and Meir , R. 2005 . S1 gene characteristics and efficacy of vaccination against infectious bronchitis virus field isolates from United States and Israel (1996 to 2000) . Avian Pathology , 34 : 194 – 203 .
- Ignjatovic , J. and Galli , L. 1994 . The S1 glycoprotein but not N or M proteins of avian infectious bronchitis virus induces protection in vaccinated chickens . Archives of Virology , 138 : 117 – 134 .
- Ignjatovic , J. and Sapats , S. 2000 . Avian infectious bronchitis virus . Revue Scientifique et technique , 19 : 493 – 508 .
- Ignjatovic , J. , Gould , G. and Sapats , S. 2006 . Isolation of a variant infectious bronchitis virus in Australia that further illustrates diversity among emerging strains . Archives of Virology , 151 : 1567 – 1585 .
- Keeler , C.L. , Reed , K.L. , Nix , W.A. and Gelb , J. 1998 . Serotype identification of avian infectious bronchitis virus by RT-PCR of the peplomer (S-1) gene . Avian Diseases , 42 : 275 – 284 .
- Kumar , S. , Tamura , K. and Nei , M. 2004 . MEGA 3: integrated software for molecular evolutionary genetics analysis and sequence alignment . Briefings in Bioinformatics , 5 : 150 – 163 .
- Kwon , H.M. and Jackwood , M.W. 1995 . Molecular cloning and sequence comparison of the S1 glycoprotein of the Gray and JMK strains of avian infectious bronchitis virus . Virus Genes , 9 : 219 – 229 .
- Kwon , H.M. , Jackwood , M.W. and Gelb , J. 1993 . Differentiation of infectious bronchitis virus serotypes using polymerase chain reaction and restriction fragment length polymorphism analysis . Avian Diseases , 37 : 194 – 202 .
- Ladman , B.S. , Loupus , A.B. and Gelb , J. 2006 . Infectious bronchitis virus S1 gene sequence comparison is a better predictor of challenge of immunity in chickens than serotyping by virus neutralization . Avian Pathology , 35 : 127 – 133 .
- Lee , C.W. , Hilt , D.A. and Jackwood , M.W. 2003 . Typing of field isolates of infectious bronchitis virus based on the sequence of the hypervariable region in the S1 gene . Journal of Veterinary Diagnostic Investigation , 15 : 344 – 348 .
- Liu , S. and Kong , X. 2004 . A new genotype of nephropathogenic infectious bronchitis virus circulating in vaccinated and non-vaccinated flocks in China . Avian Pathology , 33 : 321 – 327 .
- Liu , H.J. , Lee , L.H. , Shih , W.L. , Lin , M.Y. and Liao , M.H. 2003 . Detection of infectious bronchitis virus by multiplex polymerase chain reaction and sequence analysis . Journal of Virological Methods , 109 : 31 – 37 .
- Meulemans , G. , Boshmans , M. , Decaesstecker , M. , Van der Berg , T.P. , Denis , P. and Cavanagh , D. 2001 . Epidemiology of infectious bronchitis virus in Belgian broilers: a retrospective study, 1986 to 1995 . Avian Pathology , 30 : 411 – 421 .
- Rimondi , A. , Craig , M.I. , Vagnozzi , A. , Konig , G. , Delamer , M. and Pereda , A. 2009 . Molecular characterization of avian infectious bronchitis virus strains from outbreaks in Argentina (2001–2008) . Avian Pathology , 38 : 149 – 153 .
- Thayer , S.G. and Beard , C.W. 1998 . “ Serologic procedures ” . In A Laboratory Manual for the Isolation and Identification of Avian Pathogens , 4th edn , Edited by: Swayne , D.E. , Glisson , J.R. , Jackwood , M.W. , Pearson , J.E. and Reed , W.M. 255 – 266 . Pennsylvania, PA : The American Association of Avian Pathologists .
- Villarreal , L.Y.B. , Brandão , P.E. , Chacón , J.L.V. , Assayag , M.S. , Maiorka , P. Raffi , A.P. 2007a . Orchitis in roosters with reduced fertility associated with avian infectious bronchitis virus and avian metapneumovirus infections . Avian Diseases , 51 : 900 – 904 .
- Villarreal , L.Y.B. , Brandão , P.E. , Chacón , J.L.V. , Saidenberg , A.B.S. , Assayag , M.S. , Jones , R.C. and Ferreira , A.J.P. 2007b . Molecular characterization of Infectious bronchitis virus strains isolated from the enteric contents of Brazilian laying hens and broilers . Avian Diseases , 51 : 974 – 978 .
- Villegas , P. 1998 . “ Titration of biological suspensions ” . In A Laboratory Manual for the Isolation and Identification of Avian Pathogens , 4th edn , Edited by: Swayne , D.E. , Glisson , J.R. , Jackwood , M.W. , Pearson , J.E. and Reed , W.M. 255 – 266 . Pennsylvania, PA : The American Association of Avian Pathologists .
- Wang , C.H. and Huang , Y.C. 2000 . Relationship between serotypes and genotypes based on the hypervariable region of the S1 gene of infectious bronchitis virus . Archives of Virology , 145 : 291 – 300 .
- Worthington , K.J. , Currie , R.J.W. and Jones , R.C. 2008 . A reverse transcriptase-polymerase chain reaction survey of infectious bronchitis virus genotypes in Western Europe from 2002 to 2006 . Avian Pathology , 37 : 247 – 257 .
- Zanella , A. , Lavazza , A. , Marchi , R. , Moreno , A. and Paganelli , F. 2003 . Avian infectious bronchitis: characterization of new isolates from Italy . Avian Diseases , 47 : 180 – 185 .