Abstract
Avian astrovirus infections are widespread in many countries, and infections have been linked to enteritis and increased mortality in young poultry. Although pigeons are treated as an important poultry product in some countries, their diseases are often poorly understood and astrovirus infection in pigeons has not been reported. In the present study, faecal samples were collected during an outbreak of gastrointestinal illness in a population of Shanghai pigeons. The samples were examined for astroviruses by reverse transcription-polymerase chain reaction. Eighty-nine per cent (40/45) and 4% (2/45) were found to be positive for avian nephritis virus (ANV) and chicken astrovirus, respectively. One positive sample indicated a co-infection with both ANV and chicken astrovirus. Phylogenetic analysis based on the partial polymerase gene sequence and full-length capsid protein from published avian astrovirus sequences in GenBank revealed that the pigeon viruses detected in this study were evolutionarily closely related to chicken ANV. The present study provided evidence for the presence of astrovirus in pigeons and suggests that cross-infection between pigeons and commercial chickens was likely. Whether the astroviruses in pigeons were responsible for the diarrhoea remains to be determined.
Introduction
Astroviruses are small round, non-enveloped viruses with a diameter of 28 to 30 nm and the viral capsid contains a positive-sense RNA (Matsui & Greenberg, Citation2001). The family Astroviridae is divided into two genera of mammalian astrovirus and avian astrovirus. In poultry, astroviruses have been recognized as pathogenic agents in turkeys where infection causes diarrhoea and increased mortality. Two types of turkey astrovirus (TAstV-1 and TAstV-2) have been identified (Schultz-Cherry et al., Citation2001).
To date, two different astroviruses have been detected in chickens, avian nephritis virus (ANV) and chicken astrovirus (CAstV). ANV was considered to be the cause of subclinical pathologic changes to the kidneys of infected chickens (Frazier et al., Citation1990; Shirai et al., Citation1991). This virus was isolated and characterized in 1976 in association with mild growth depression, kidney lesions, and mortality in young chickens (Yamaguchi et al., Citation1979) and was originally regarded as a picornavirus. In 2000, however, ANV was reclassified as a member of the genus Avastrovirus of the family Astroviridae (Imada et al., Citation2000). Studies undertaken in Japan have shown that there are at least two different ANV serotypes (Shirai et al., Citation1992). The CAstV virus was isolated from broiler chicks with runting/stunting syndrome in the Netherlands; CAstV shares some sequence identity with ANV but is serologically distinct from it (Baxendale & Mebatsion, Citation2004).
The astrovirus RNA genome is 6.8 to 7.9 kb in length. It contains three open reading frames (ORFs) denoted ORF1a, ORF1b, and ORF2 that encode a serine protease, an RNA-dependent RNA polymerase (RdRP) and a capsid precursor protein, respectively (Imada et al., Citation2000).
Astroviruses have been linked to enteritis and increased mortality in young turkeys, chickens, and guinea fowl, nephritis in chickens, and a fatal hepatitis in ducklings (Yamaguchi et al., Citation1979; Imada et al., Citation1980; McNulty et al., Citation1989; Shirai et al., Citation1990; Cattoli et al., Citation2007). Severity of astrovirus infections ranges from subclinical infection to death, and young chicks are most susceptible.
In some countries pigeons are considered important poultry, but it is not known if they can be infected by the astrovirus or if infection causes serious disease. The aims of the present study were to look for the presence of astroviruses by reverse transcriptase-polymerase chain reaction (RT-PCR) in diarrhoea samples from young pigeons (<2 weeks of age) and to determine the phylogenetic relationship between a putative pigeon astrovirus and other avian astroviruses.
Materials and Methods
Sampling
A total of 45 faecal samples from pigeons younger than 2 weeks of age were collected from the Animal Center for D isease Control in Shanghai, China in September 2010 during an outbreak of gastrointestinal illness. In addition, 30 faecal samples from healthy pigeons (<2 weeks old) were collected at specialized pigeon centres.
Extraction of total RNA
Faecal samples were diluted in phosphate-buffered saline at a ratio of 1:6 (wt/vol.) and homogenized. Total nucleic acid was extracted from 200 µl each sample by TRIzol Reagent (Invitrogen Inc., Carlsbad, California, USA) according to the manufacturer's instructions. Total RNA was eluted in 30 µl diethyl pyrocarbonate (DEPC)-treated water and immediately stored at –80°C until analysis.
RT-PCR detection pigeon astrovirus
Two sets of primers were used for detecting astroviruses
The first set was designed to amplify a 450 base pair (bp) product from ANV. The degenerate primers PV-F (5′-TAGAATCACTCTTCTTCCAACTGGA-3′) and PV-R (5′-TCAAGGTCTGGCAGCCTGCGC-3′) were targeted to a conserved region located in the ORF1b gene (3936 to 4385 bp of ANV G-4260 accession number AB033998). The second set of primers (CAstV-for and CAstV-rev) were designed by Smyth et al. (Citation2009) to detect CAstV by amplifying a 510 bp region of the ORF1b gene. Reverse transcription was performed using the PrimeScript 1st Strand cDNA Synthesis Kit (TaKaRa Bio Inc., Dalian city, Japan) according to the manufacturer's protocol. The cDNA template (5 µl) was used in a 50 µl reaction mixture and amplified with PCR TaqMix (TaKaRa Bio Inc., Dalian city, Japan). The amplification programme consisted of an initial 5 min denaturation step at 94°C, followed by 40 cycles of 94°C for 30 sec, 55°C for 30 sec, and 72°C for 90 sec, followed by a final elongation step at 72°C for 7 min. As a positive control, we included a chicken faecal sample previously shown to be positive for astrovirus by RT-PCR.
According to the GenBank sequence of ANV (accession numbers AB033998 and HM029238), we designed a pair of primers towards the ORF2 of ANV to amplified pigeon ANV (PeANV10 sample) capsid protein: PORF-F, 5′-ATGCCTGGCCCTGCCGGCCCTGGTA-3′; and PORF-R, 5′-TTAGATAGTAAAGCGCTTTGCGTCG-3′.
The RT reaction was performed with the primer AV12 (5′-TTTTTTTTTTTTTTTTTTGC-3′), which has the sequence within the 3′ terminal of the complete astrovirus genome. Reverse transcription was performed using the PrimeScript 1st Strand cDNA Synthesis Kit (TaKaRa Bio Inc.) according to the manufacturer's protocol. The PCR was performed using the TransTaq High Fidelity (HiFi) PCR superMix II (TransGen Biotech, Beijing, China) based on the system's protocol, which was conducted with 5 µl cDNA, 25 µl TransTaq HiFi PCR SuperMix, 1 µl PORF-F and PORF-R (25 µM each), and distilled water to a total volume of 50 µl and amplified as follows: 94 °C for 5 min; 45 cycles of PCR (94°C for 30 sec, 55°C for 30 sec, 70°C for 3 min) and a final 15-min extension at 72°C.
Sequence analysis and cloning
All of the RT-PCR amplified DNA products were analysed by agarose gel electrophoresis, and amplicons of the correct size were excised, extracted with the AxyPrep DNA gel extraction kit (Axygen Inc., California, USA), cloned into PMD-18T (TaKaRa Bio Inc., Dalian city, Japan) and sequenced (Shanghai Sangon, Shanghai, China).
All sequences were compared using the National Center for Biotechnology information BLAST program (http://www.ncbi.nlm.nih.gov/blast.cgi) and entered into the Mega 4 program (http://www.megasoftware.net/) to build phylogenetic trees using the neighbour-joining method. Segments of the partial polymerase gene and capsid protein from published avian astrovirus sequences in GenBank were included in the phylogenetic analysis.
GenBank accession
The nucleotide sequences determined in this study were deposited in GenBank (http://www.ncbi.nlm.nih.gov/Genbank/idex.html) and assigned the accession numbers HQ662572 to HQ662579. The pigeon ANV virus caspid protein sequence was assigned accession number JF422783.
Results
Detection of astrovirus cDNA in pigeon faecal samples
ANV transcripts of approximately 450 bp were amplified using primers PV-F and PV-R. From the 45 pigeon diarrhoea samples, 40 (89%) were positive for ANV. Application of the primers CAstV-for and CAstV-rev produced fragments of approximately 510 bp from two samples. In contrast, of 30 stool samples from healthy pigeons, none was positive for either ANV or CAstV. In the positive samples, only one sample was positive for both ANV and CAstV. Following cloning and sequencing of the RT-PCR products, BLAST search analyses with the predicted amino acid sequences revealed that the isolate ANV in pigeons was most closely related to ANV G-4260 (AB033998), and the CAstV-like isolate in pigeons was most closely related to the chicken astrovirus isolated by Smyth et al. (Citation2009).
Sequence analyses of pigeon ANV
Of the 40 positive samples amplified by the PV-F and PV-R primers, six positive field sample sequences were determined. The nucleotide sequences of the amplicons were deposited in GenBank under accession numbers HQ662572 to HQ662577. The sequences were aligned to each other, and nucleotide identity levels varied between 89 and 99%, while predicted amino acid homology was 94 to 99%.
Owing to the limited sequence information available in public databases, a 208-nucleotide region of ORF1b (from 3936 to 4143 by numbering of the ANV G-4260 genome) was compared with other ANV strains. The nucleotide sequence alignments indicated 83 to 99% identity between the ANVs detected in pigeons (PeANV) and the reference ANV strain (accession numbers DQ324827 to DQ324838), 84 to 86% for strain AB033998, and 85 to 86% for strain EU669002. The highest amino acid identity values (96 to 97%) were shared by PeANV and HM029238 (Chinese ANV strain). Similarly, these PeANV strains showed highest identity (93 to 99%) with the nucleotide sequence in BLAST searches ().
Table 1. Nucleic acid identity (%) among pigeon astrovirus isolates with other avian astroviruses.a
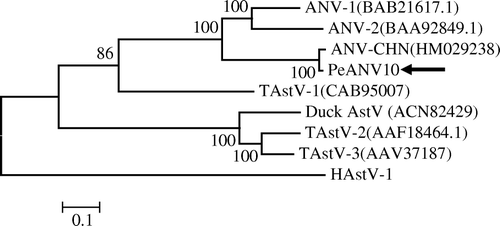
In an effort to determine the phylogenetic relationship between PeANV and other ANV strains, a phylogram was constructed from a 208 bp fragment located in the RdRP gene (a). For the construction of the tree, turkey astrovirus (TAstV) and duck astrovirus (DAstV) were used as the outgroups. Examination revealed that the TAstV, DAstV, and ANV formed two major groups. Within the ANV group, isolates representing six pigeon astrovirus sequences were closely related to one another, with 93 to 99% nucleotide identity. PeANV18, PeANV19, PeANV40 PeANV13 and PeANV35 were more closely related to the ANV-China strain (HM029238) and PeANV10 was more closely related to ANV USA strains (DQ324827). All of the isolated pigeon ANV sequences were more closely related to ANV USA strains (DQ324827 to DQ324838) than to the ANV-G4260 Japan (AB033998) or EF91-276 Northern Ireland (EU669002) strains.
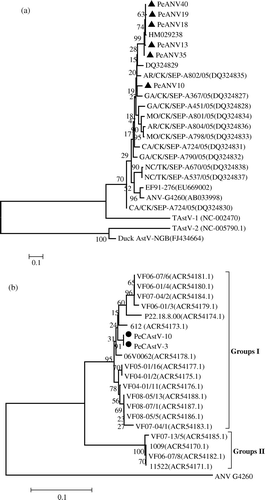
Figure 1. Phylogenetic relationship of pigeon astroviruses based on the sequence of overlapping regions of selected field and reference samples. The tree was constructed with Mega 4 using neighbour-joining and 1000 bootstrap replicates (bootstrap values are shown). GenBank accession numbers presented in parentheses. 1a: Phylogenetic trees based on the ORF1b nucleotide sequence (208 nucleotides) of ANV (3936 to 4143 nucleotides, AB033998). Black triangles indicate pigeon ANV (PeANV). The reference turkey and duck astrovirus was used as the outgroup. 1b: Phylogenetic trees based on the ORF1b amino acid sequence (140 amino acids) from selected field samples and isolates of chicken astrovirus. Black spheres indicate pigeon astrovirus (PeCAstV). The reference avian nephritis virus (G-4260) was used as the outgroup.
As expected, with application of primers PORF-F and PORF-R, the complete PeANV10 ORF2 gene sequence was obtained (accession number JF422783). The PeANV10 full-length ORF2 sequence was 2043 bp. When we compared the entire region sequence of ORF2 with that of reference strains of ANV, the identities shared by PeANV10 with G-4260 was relatively low (nucleotides 66% and amino acids 59%, respectively) in comparison with the identities shared by the Chinese ANV strain (ANV-CHN) (nucleotides 95% and amino acids 97%, respectively).
Phylogenetic analyses based on amino acid sequences of the ORF2 region by the neighbour-joining method showed that PeANV10 grouped within the ANV clade and was most closely related to ANV-CHN ().
Sequence analyses of pigeon CAstV
Two faecal samples were positive for CAstV using the primers CAstV-for and CAstV-rev. The nucleotide sequences of the amplicons (510 bp) were determined and have been deposited in GenBank with accession numbers HQ662578 to HQ662579. A phylogenetic tree was constructed based on the ORF1b amino acid sequence (140 amino acids) from selected field samples and isolates of the CAstV overlapping region (b). Based on the partial CAstV ORF1b amino acid sequence phylogenetic analysis of earlier studies, two CAstV groups (Groups I and II) could be distinguished (Pantin-Jackwood et al., Citation2006; Smyth et al., Citation2009; Todd et al., Citation2009). Pigeon CAstV (PeCAstV) ORF1b sequences closely similar to the 612 isolates were assigned to Group I. They shared 95 to 97% amino acid identity with the 612-like CAstVs and displayed 82 to 83% amino acid identity with Group II CAstVs. Overall, the PeCAstVs showed 88 to 92% nucleotide identity over the 420 nucleotide ORF1b sequences compared with Group I ().
Discussion
This is the first paper to report astrovirus infection in pigeons with diarrhoea. Although earlier investigations indicated that chickens and turkeys were infected with astrovirus, there was uncertainty as to whether wild birds like those studied also carried an astrovirus. In the present paper, the pigeons that provided the diarrhoea samples belonged to the same age group, from 5 days old to around 2 weeks old. Investigations of the samples revealed the presence of ANV in 89% and CAstV in 4% of the diarrhoea faecal samples. Mandoki et al. (Citation2006) confirmed the presence of ANV in 69% of chickens with diarrhoea and growth retardation examined in Hungary. Day et al. (Citation2007) reported ANVs in enteric samples from 56% of sick and healthy chickens originating in the USA. The ANV infection rate of 89% in our study suggests that ANV may also infect free-living pigeon flocks but investigation of more pigeon faecal samples from varied sources needs to be undertaken to establish pigeon astrovirus prevalence in China.
The established data suggest that astroviruses are the most frequently identified viruses in turkey and chicken flocks. In the present paper, sequence comparisons of the pigeon ANV partial RdRP gene and full-length ORF2 gene with other avian astroviruses confirmed that pigeon ANV shared high nucleotide and amino acid identity with ANV. Phylogenetic analyses revealed that pigeon ANV was most closely related to a Chinese ANV strain (ANV-CHN), which was isolated from layer faecal samples in Sichuan province, China by our laboratory in 2009. The high identity of structural protein nucleotide and amino acid sequences showed that pigeon ANV and ANV-CHN were probably from the same virus. These findings are of interest because they suggest that cross-infections may take place between pigeons and commercial chickens. However, it remains to be determined whether the described pigeon ANV originated from chicken ANV.
The chicken astrovirus CAstV was identified by Baxendale & Mebatsion (Citation2004) and later detected by Pantin-Jackwood et al. (Citation2006) in both problem and healthy US flocks. In our study, we detected two samples positive for CAstV. Interestingly, one sample showed co-infection of ANV and CAstV. This indicates that more than one species of astrovirus was circulating in these pigeon flocks. Similarly TAstV-1, TAstV-2 and ANV were detected simultaneously in chickens on the same farm (Pantin-Jackwood et al., Citation2006). Multiple astroviruses were the most frequently identified combination in both diseased and healthy flocks; however, the significance of ANV and CAstV co-infection in our pigeon sample is not known and further characterization is needed.
There are few specialized pigeon centres and limited publications in the scientific literature, so diseases of pigeons are often poorly understood by the veterinarian. To our knowledge, this is the first demonstration of astrovirus infection in a pigeon flock during a diarrhoea outbreak. Several research groups have examined astrovirus infection and pathology in chicks, but the pathogenesis of the signs is poorly understood. Whether the astroviruses detected in pigeons contributed to the outbreak of diarrhoea remains to be determined. The pathogenic properties of astroviruses in pigeons must be characterized to further understand astrovirus infection in pigeon flocks and the relationship between astroviruses in pigeons and other ANV and CAstV viruses.
Acknowledgements
The authors thank Dr Congli Yuan for advice on the statistical analyses and excellent technical assistance. The pigeon diarrhoea samples were kindly provided by Animal Center for Disease Control, Shanghai, China.
References
- Baxendale , W. and Mebatsion , T. 2004 . The isolation and characterization of astroviruses from chickens . Avian Pathology , 33 : 364 – 370 .
- Cattoli , G. , De Battisti , C. , Toffan , A. , Salviato , A. , Lavazza , A. , Cerioli , M. and Capua , I. 2007 . Co-circulation of distinct genetic lineages of astroviruses in turkeys and guinea fowl . Archives of Virology , 152 : 595 – 602 .
- Day , J.M. , Spackman , E. and Pantin-Jackwood , M. 2007 . A multiplex RT-PCR test for the differential identification of turkey astrovirus type 1, turkey astrovirus type 2, chicken astrovirus, avian nephritis virus, and avian rotavirus . Avian Diseases , 51 : 681 – 684 .
- Frazier , J.A. , Howes , K. , Reece , R.L. , Kidd , A.W. and Cavanagh , D. 1990 . Isolation of noncytopathic viruses implicated in the aetiology of nephritis and baby chick nephropathy and serologically related to avian nephritis virus . Avian Pathology , 19 : 139 – 160 .
- Imada , T. , Yamaguchi , S. , Mase , M. , Tsukamoto , K. , Kubo , M. and Morooka , A. 2000 . Avian nephritis virus (ANV) as a new member of the family Astroviridae and construction of infectious ANV cDNA . Journal of Virology , 74 : 8487 – 8493 .
- Imada , T. , Yamaguchi , S. , Mura , N. and Kawamura , H. 1980 . Antibody survey against avian nephritis virus among chickens in Japan . National Institute of Animal Health Quarterly , 20 : 79 – 80 .
- Mandoki , M. , Bakonyi , T. , Ivanics , E. , Nemes , C. , Dobos-Kovacs , M. and Rusvai , M. 2006 . Phylogenetic diversity of avian nephritis virus in Hungarian chicken flocks . Avian Pathology , 35 : 224 – 229 .
- Matsui , S.M. and Greenberg , H.B. 2001 . Astroviruses , in D.M. Knipe , P.M. Howley Fields Virology , 4th edn, Vol. 1 . Baltimore, MD Lippincott Williams and Wilkins , pp. 875 – 893 .
- McNulty , M.S. , Connor , T.J. and McNeilly , F. 1989 . A survey of specific pathogen-free chicken flocks for antibodies to chicken anaemia agent, avian nephritis virus and group A rotavirus . Avian Pathology , 18 : 215 – 220 .
- Pantin-Jackwood , M.J. , Spackman , E. and Woolcock , P.R. 2006 . Molecular characterization and typing of chicken and turkey astroviruses circ ulating in the United States: implications for diagnostics . Avian Diseases , 50 : 397 – 404 .
- Schultz-Cherry , S. , King , D.J. and Koci , M.D. 2001 . Inactivation of an astrovirus associated with poult enteritis mortality syndrome . Avian Diseases , 45 : 76 – 82 .
- Shirai , J. , Nakamura , K. , Shinohara , K. and Kawamura , H. 1991 . Pathogenicity and antigenicity of avian nephritis isolates . Avian Diseases , 35 : 49 – 54 .
- Shirai , J. , Obata , H. , Nakamura , K. , Furuta , K. , Hihara , H. and Kawamura , H. 1990 . Experimental infection in specific-pathogen-free chicks with avian reovirus and avian nephritis virus isolated from broiler chicks showing runting syndrome . Avian Diseases , 34 : 295 – 303 .
- Shirai , J. , Tanimura , N. , Uramoto , K. , Narita , M. , Nakamura , K. and Kawamura , H. 1992 . Pathologically and serologically different avian nephritis virus isolates implicated in etiology of baby chick nephropathy . Avian Diseases , 36 : 369 – 377 .
- Smyth , V.J. , Jewhurst , H.L. , Adair , B.M. and Todd , D. 2009 . Detection of chicken astrovirus by reverse transcriptase-polymerase chain reaction . Avian Pathology , 38 : 293 – 299 .
- Todd , D. , Wilkinson , D.S. , Jewhurst , H.L. , Wylie , M. , Gordon , A.W. and Adair , B.M. 2009 . A seroprevalence investigation of chicken astrovirus infections . Avian Pathology , 38 : 301 – 309 .
- Yamaguchi , S. , Imada , T. and Kawamura , H. 1979 . Characterization of a picornavirus isolated from broiler chicks . Avian Diseases , 23 : 571 – 581 .