Abstract
The complete capsid gene sequences of 24 chicken astroviruses (CAstVs), collected in the UK, Germany, the Netherlands and South Africa from the 1980s to 2008, were determined and compared with that of a US CAstV (UGA-2006). Pairwise comparisons and phylogenetic analysis demonstrated the existence of two major capsid groups, designated A and B, which shared 38 to 40% amino acid identity. CAstVs from groups A and B shared capsid protein identities ranging from 26 to 38% with other avian astroviruses. The group A CAstVs comprised three subgroups, which displayed inter-subgroup identities ranging from 77 to 82%, while group B comprised two clearly separated subgroups, Bi and Bii, which displayed intra-subgroup identities of 97 to 99% and 94 to 99%, respectively, and shared inter-subgroup identities of 84 to 85%. Phylogenetic analyses performed with contiguous open reading frame 1b (polymerase) and open reading frame 2 (capsid) CAstV sequences showed that CAstVs from capsid subgroup Bi had polymerase genes that differed from those possessed by CAstVs belonging to group A and subgroup Bii. The N-terminal capsid regions (residues 1 to 415) were more conserved than the C-terminal regions, with the C-terminal regions of the subgroup Bi and Bii CAstVs sharing 76 to 78% amino acid identity, while the C-terminal regions of the A subgroups displayed identities less than 75%. CAstVs representative of both capsid groups and more than one subgroup were detected within the same broiler flock. The high level of capsid sequence diversity observed in this study has important implications for both the control and diagnosis of CAstV infections.
Introduction
To date, two different astroviruses have been detected in chickens. Avian nephritis virus (ANV), originally regarded as a picornavirus, was characterized as the first astrovirus of chickens in 2000 (Imada et al., Citation2000). Baxendale & Mebatsion (Citation2004) described the characterization of a novel astrovirus, named chicken astrovirus (CAstV), which was detected in broiler chicks with runting problems in the Netherlands. In common with other avian and mammalian astroviruses, ANV and CAstV possess single-stranded, positive-sense RNA genomes (~7 kb in size) that encompass three open reading frames (ORFs). ORF 1a, located close to the 5′ terminus of the genome, encodes the viral protease and is followed by ORF 1b, which encodes the RNA polymerase. ORF 2, encoding the precursor capsid protein, is located downstream of ORF 1b and prior to the 3′-untranslated region of the genome. Earlier studies have shown that ANV and CAstV are antigenically distinct and share relatively low levels of ORF 1b and ORF 2 sequence identity (McNulty et al., Citation1991; Todd et al., Citation2009a, Citation2011).
CAstVs, which shared high levels of ORF 1b nucleotide sequence identity with the first characterized CAstV (Baxendale & Mebatsion, Citation2004), hereafter known as the reference CAstV, have also been detected in US broiler chicken flocks affected by growth problems (Pantin-Jackwood et al., Citation2006). More recently, we have reported the identification of antigenically different CAstV isolates, including the FP3, 612 and 11672 isolates, which were originally described as enterovirus-like viruses (Todd et al., Citation2009a). CAstVFP3 was isolated from dead-in-shell chicks during an investigation of early broiler mortality in the UK (Spackman et al., Citation1984), while CAstV612 was isolated from South African broiler chickens with respiratory problems (McNeilly et al., Citation1994). CAstV11672 was isolated from 1-day-old chicks as part of an investigation into broiler hatchability problems (Todd et al., Citation2009a). Whereas isolate 612 shared close antigenic and sequence similarities with the reference CAstV, isolates FP3 and 11672 were closely related to one another and more distantly related to the reference CAstV and 612 isolates in terms of antigenicity and partial ORF 1b sequence (Todd et al., Citation2009a).
Our interest in CAstVs relates to their possible involvement in causing enteritis and growth depression in chickens (Guy et al., Citation2008). Recently developed reverse transcription polymerase chain reaction (RT-PCR) tests have shown that CAstVs are commonly detected in intestinal samples from UK and US broiler flocks affected by enteritis and/or growth retardation (Pantin-Jackwood et al., Citation2006; Smyth et al., Citation2009, 2010). Experimental infections of 1-day-old specific pathogen free (SPF) and broiler chicks have demonstrated that CAstVs can cause varying degrees of growth retardation (McNeilly et al., Citation1994; Smyth et al., Citation2007; Todd et al., unpublished results 2011). The study by Smyth et al. (Citation2007) also showed that following experimental infection of SPF chicks with the FP3 isolate, histological lesions and virus-specific antigen were detected in the intestine, kidney and pancreas, indicating that, like ANV, CAstVs have the ability to infect internal organs. Subsequent work has shown that CAstVs were detected in high proportions of kidney as well as intestinal content samples that were obtained in longitudinal surveys of four broiler flocks (Smyth et al., Citation2010).
Serological investigations have also demonstrated that CAstV infections are highly prevalent (McNeilly et al., Citation1994; Baxendale & Mebatsion, Citation2004; Todd et al., Citation2009b). In the study by Todd et al. (Citation2009b), separate indirect immunofluorescence (IIF) tests were developed and applied for the detection of serum antibodies reactive with the 612 and 11672 CAstV isolates, which display very low levels of antigenic cross-reactivity by IIF. That investigation showed that infections with both CAstVs were very common in UK broiler and broiler breeder flocks, were widespread in poultry organizations within Europe, and were detected at lower levels in grandparent and great-grandparent flocks.
To date, comparatively few sequence data exist for the CAstV. Working with partial ORF 1b sequences determined for 20 CAstVs present in UK samples, Smyth et al. (Citation2009) described the existence of two polymerase groups, named I and II. These shared inter-group nucleotide identities in the range 76 to 79%, with the intra-group nucleotide identities being 85 to 99% and 95 to 99% for the Group I and Group II CAstVs, respectively (Smyth et al., Citation2009). More recently, Pantin-Jackwood et al. (Citation2011) have reported that the partial ORF 2 (capsid) sequences of six CAstVs from US chickens displayed nucleotide and amino acid identities of 74 to 92% and 85 to 98%, respectively.
On the basis of its probable interactions with the host's immune system and host cell receptors, the astrovirus capsid protein is likely to be the major determinant of virus antigenicity and cell tropism. As such, it will influence pathogenesis and pathogenicity. In this paper we report the capsid protein gene sequences and sequence comparisons of 25 CAstVs. These included six CAstV isolates, which can be propagated in chick embryos or cell culture, as well as CAstVs that were detected by RT-PCR in samples collected from UK broiler flocks affected by growth retardation problems in 2008. Because CAstVs are relatively difficult to isolate and to characterize antigenically, capsid gene sequence determination provides a useful indicator of possible antigenic relatedness.
Materials and Methods
Virus isolates and virus growth
For the purposes of this paper, virus isolates are regarded as viruses that have been propagated in chick embryos or in cell culture and are distinct from unpropagated viruses that are present in field samples. Information relating to the source and propagation in SPF embryonated eggs of the 11672, 612, FP3 and 11522 CAstV isolates was described previously (Todd et al., Citation2009a). CAstV1010 was also grown in SPF eggs following its isolation from weak chicks, which were submitted as part of an investigation into hatchability problems that occurred in the UK in 2004 and 2005 (Todd, 2005 unpublished results). Virus pools produced as 10% homogenates of whole embryos or embryo livers were used for RNA extraction.
CAstVVF08-29 was isolated from a pool of intestinal contents from 4-day-old SPF chicks that had been experimentally infected with pooled intestinal contents collected from 5-day-old and 7-day-old broiler chickens, which were collected in a longitudinal survey undertaken in 2008 (Todd et al., Citation2010). This virus was first propagated in SPF embryonated eggs following yolk sac inoculation and then adapted to grow in primary chick embryo liver cells and then LMH cells. The P22.18.8.00 isolate of CAstV, which we have designated as the “reference” CAstV, was kindly provided by Dr Ian Tarpey (Intervet-Schering Plough, Milton Keynes, UK).
Sample origin and processing
Intestinal contents were obtained from broiler chickens, reared in the UK and Germany, and stored at −80°C. Intestinal contents were processed as 10% suspensions in phosphate-buffered saline as described in Smyth et al. (Citation2009). The supernatants were collected and transferred to fresh tubes for storage at −80°C until required. Samples from the UK were kindly provided by A. Fernadez-Gutierrez (Aviagen Ltd, Midlothian, UK), M. Alcorn (St Davids Poultry Team Ltd, Exmouth, UK), M. Hardy (St Davids Poultry Team Ltd, Dungannon, Northern Ireland), and C. Prins, G. Hayes and A. Walker (Slate Hall Veterinary Practice Ltd, Cambridge, UK). Eighty pooled samples were collected in longitudinal surveys of four broiler flocks as described previously (Todd et al., Citation2010). Briefly, intestinal contents from ~12 birds were sampled from each flock at days 0, 4 or 5, 7, 10, 14, 21, 28 and 35 days (, organization D). The samples were grouped into four pools at each timepoint and processed by homogenization as described above. Dr P. Otto (Friedrich Loeffler Institute, Federal Institute for Animal Health, Jena, Germany) provided sample 05V150/152/154, which is a filtrate prepared from field samples from three birds.
Table 1. Origin of CAstV capsid sequences.
RNA extraction
Viral RNA was extracted from 140 µl of each supernatant using the QIAamp Viral RNA Mini Kit (Qiagen, Crawley, UK) according to the manufacturer's instructions. Each RNA was eluted in 30 µl RNase-free water.
RT-PCR assay
The forward primer (CAstV A For, 5′-AGC CTC AAA GTA TAA GAC GCAG-3′) based in ORF 1b was used with a composite, anchored oligo-dT reverse primer to generate a fragment of ~3.22 kb from CAstV11672 and CAstVFP3 RNA extracts (), by one-step RT-PCR, which was performed on the RNAs using the SuperScript III One-Step RT-PCR System with Platinum® Taq DNA Polymerase kit (Invitrogen, Paisley, UK). Each reaction contained 1×reaction buffer, 1 µM each primer, 1 µl enzyme mix, 2.5 µl RNA and diethyl pyrocarbonate-treated water to 25 µl. Amplification occurred in a Veriti thermocycler (Applied Biosystems, Paisley, UK) starting with a reverse transcription step of 45°C for 30 min, then an initial denaturing step of 95°C for 2 min, followed by 40 cycles of denaturation at 95°C for 30 sec, and primer annealing at a temperature of 45°C for 30 sec and an extension time of 3.5 min. There was a final extension step at 68°C for 7 min. These RT-PCR fragments were gel-excised, purified, cloned and sequenced as described below. From the sequences of these 3.22 kb fragments, two pairs of primers were designed to amplify (i) a fragment of ~3.2 kb largely similar to but slightly smaller than the 3.22kb fragment, and (ii) a fragment of ~2.2 kb ORF 2 product from CAstVs (). For (i) the primer sequences were 3.2 CAstV For (5′-AGC CTC AAA GTA TAA GAC GC-3′) and 3.2 CAstV Rev (5′-GCC AAT TAA TCT AAT TCG AAA TC-3′), and these were used in an RT-PCR reaction as described above but with an annealing temperature of 60°C. For (ii) the forward primer, located in the intergenic region between ORFs 1b and 2 (), was CAstV_PRECAP (5′-TAG AGG GAT GGA CCG AAA TAT AGC AGC-3′), while the reverse primer, CAstV_POSTCAP (5′-TGCAGCTGTACCCTCGATCCTA-3′), was located immediately after the end of the capsid gene. This primer set was used in RT-PCR reactions as described above but with an annealing temperature of 50°C and an extension time of 2.5 min. An additional primer set was used to amplify a fragment of ~900 base pairs (bp), which spanned the 3′ region of ORF 1b, the intergenic region and the 5′ region of ORF 2 () from selected CAstVs. The forward primer was the CAstV-for degenerate primer described in Smyth et al. (Citation2009) and this was used in combination with reverse primers that were specific for each of the viruses sequenced. The RT-PCR products were electrophoresed at 125 V for 40 min on a 1% agarose gel in 1×Tris–acetate ethylenediamine tetraacetic acid buffer and were visualized by ethidium bromide staining and ultraviolet transillumination. Amplicons of the appropriate sizes were excised from the gel and purified using the Wizard SV Gel and PCR Clean-Up System (Promega, Southampton, UK).
Figure 1. The 3′ region of the CAstV genome showing the locations of the amplicons produced by RT-PCR for cloning and sequencing. The genomic regions encompassing part of ORF 1b, ORF 2 and the 3′ untranslated region (3′ UTR) are shown together with the locations of the intergenic region (*) and poly A tail (▪). Arrows indicate forward and reverse primers, and dotted lines represent the regions amplified by RT-PCR, together with their sizes (3.22 kb, 3.2 kb, 2.2 kb and 900 bp).

Cloning and sequence analysis
Gel-purified RT-PCR fragments of approximately 3.22 kb, 3.2 kb and 2.2kb were cloned into the pCR2.1®TOPO vector (Invitrogen). The cloned inserts and gel-purified 900 bp PCR amplicons were sequenced in both directions using the BigDye® Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems), the M13 vector primers and the internal sequencing primers designed by primer walking. The products were sequenced commercially. Sequencing data were analysed using the Vector NTI suite (Invitrogen). A multiple alignment was constructed using the Clustal W2 program (Larkin et al., Citation2007) available on the European Bioinformatics Institute website (http://www.ebi.ac.uk/Tools/clustalw2/). Phylogenetic analysis was conducted using MEGA version 4 (Tamura et al., Citation2007).
Accession numbers
The Genbank accession numbers of the other avian astroviruses used in this investigation are AB033998 (ANV serotype 1 [ANV-1]), AB046864 (ANV serotype 2 [ANV-2]), Y15936 (turkey astrovirus type 1 [TAstV-1]), AF206663 (turkey astrovirus type 2 [TAstV-2]) and FJ434664 (duck astrovirus [DAstV]). The accession numbers of CAstV capsid sequences generated within this study are JN582305 (11522), JN582306 (1010), JN582307 (VF06-1/1), JN582308 (VF06-1/2), JN582309 (VF06-1/4), JN582310 (VF06-7/5), JN582311 (VF06-7/8), JN582312 (05V150/152/154), JN582313 (VF06-7/3), JN582314 (VF07-4/2), JN582315 (VF08-29), JN582316 (VF08-3), JN582317 (612), JN582318 (P22.18.8.00), JN582319 (VF08-56), JN582320 (VF08-60), JN582321 (VF08-46), JN582322 (VF08-65), JN582323 (VF08-54), JN582324 (VF08-18/7), JN582325 (VF08-36), JN582326 (VF08-48), JN582327 (11672), and JN582328 (FP3). The capsid sequence of the UGA-2006 CAstV (Kang et al., Citation2012) was kindly provided by Dr Egbert Mundt (College of Veterinary Medicine, University of Georgia, Athens, Georgia, USA).
Results
Amplification, cloning and sequencing of ORF 2-containing and ORF 1b-containing cDNA fragments
RT-PCR using an ORF 1b-based forward primer (CAstV A For) and the anchored oligo-dT primers successfully amplified a cDNA fragment of 3.22 kb from RNA extracted from embryo-grown CAstV11672 and CAstVFP3 isolates. Sequencing analysis of the cloned fragments confirmed that these were CAstV specific, each comprising a region of 730 nucleotides at the 3′ end of ORF 1b, a 24-nucleotide intergenic region, the complete CAstV ORF 2 (2217 nucleotides) and the 3′-untranslated region (~280 nucleotides) (). Fragments of either ~3.2 kb (VF06-1/1, VF06-1/2, VF06-1/4 and VF06-7/5) or ~2.2 kb (ORF 2 only) were amplified from the remaining CAstV isolates and intestinal content samples that were known to contain CAstV RNA. These were cloned and sequenced. Evidence of contiguous ORF 1b and ORF 2 sequences was further obtained by sequencing fragments of ~900 bp, which spanned the 3′ regions of ORF 1b and 5′ regions of ORF 2 from eight CAstV samples (1010, VF08-48, 05V150/152/154, VF08-54, 612, P22-18.8.00, VF07-7/3, VF07-4/2).
Pairwise comparison of the ORF 2 sequences
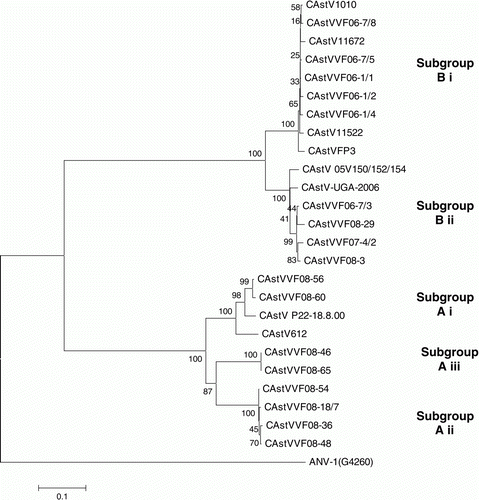
The 25 CAstV ORF 2 sequences, including that sequenced elsewhere (CAstV-UGA-2006), ranged from 2154 to 2229 nucleotides and encoded capsid proteins that ranged from 718 to 743 amino acids. A multiple alignment of the 25 capsid sequences was undertaken and pairwise identities obtained. Two major groups, designated A and B, were recognized, with group A comprising three subgroups (Ai, Aii and Aiii) and group B comprising two distinct subgroups (Bi and Bii) (). The capsids comprising group A ranged from 718 to 721 amino acids, whereas the capsids of subgroups Bi and Bii comprised 738 and 743 amino acids, respectively. Eight field samples and two of the isolates, including the reference CAstV (P22.18.00), clustered together in group A and shared nucleotide and amino acid identities of 71 to 100% and 77 to 100%, respectively. Five field samples and four of the isolates clustered tightly in subgroup Bi and shared nucleotide and amino acid identities of 91 to 99% and 97 to 99%, respectively. Subgroup Bii, comprising five field samples and one isolate (VF8-29), shared nucleotide and amino acid identities of 87 to 99% and 94 to 99%. The nucleotide and amino acid identities shared by the CAstVs of groups A and B were low at 49 to 54% and 38 to 40%, respectively. The amino acid identities shared by CAstVs belonging to subgroups Bi and Bii were 84 to 85%, while the inter-subgroup amino acid identities shared by the A subgroups were 77 to 78% (Ai and Aii), 78 to 80% (Ai and Aiii) and 82% (Aii and Aiii) ().
Table 2. Pairwise comparison of CAstV subgroups showing amino acid (nucleotide) values.
Pairwise comparisons of the CAstV capsid sequences with those of other avian astroviruses showed that the CAstV612 from group A shared amino acid identities of 28% with ANV-1, 26% with ANV-2, and 38% with TAstV-1, TAstV-2 and DAstV, while the group B CAstV11672 shared identities of 27% with ANV-1 and ANV-2, and 35% with TAstV-1, TAstV-2 and DAstV.
Phylogenetic analyses
A phylogenetic tree was constructed by comparing the deduced capsid protein sequences from 25 CAstVs using the Mega 4 program and the neighbour-joining algorithm with 1000 bootstrap replicates (). The phylogenetic distribution was consistent with the results obtained by pairwise comparison in that two major clusters were evident, corresponding to groups A and B. Group B comprised two clearly separated subclusters, which corresponded to subgroups Bi and Bii. The nine CAstVs comprising group A were distributed in three minor subclusters, which corresponded to subgroups Ai, Aii and Aiii.
Figure 2. Phylogenetic tree of CAstVs based on capsid amino acid sequences. The tree was constructed using Mega 4 (Tamura et al., Citation2007) using the neighbour-joining method and 1000 bootstrap replicates (bootstrap values are shown on the tree). Data relating to the origin of the CAstVs, including the Genbank accession numbers, are shown in . ANV-1 was used to root the tree.
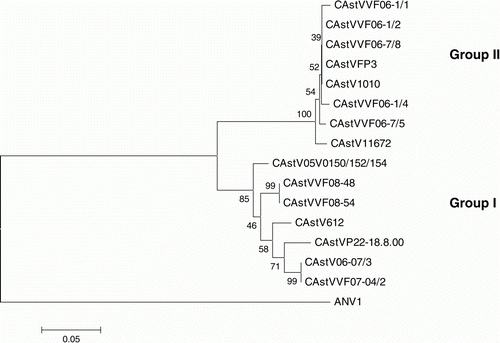
A second phylogenetic tree was similarly constructed by comparing partial 3′ ORF 1b amino acid sequences derived from 15 of the 25 CAstVs that had been compared in terms of their ORF 2 sequences (). Sequence determination and analysis showed that, in each case, the partial ORF 1b sequence had been contiguous with the ORF 2 sequences compared in . The 15 CAstVs were distributed in two major clusters, which we have designated polymerase groups I and II. Examination of the analysed CAstVs indicated that all eight CAstVs comprising polymerase group II possessed capsid sequences that were assigned to capsid subgroup Bi (), whereas those CAstVs comprising polymerase group I had capsids that were assigned to capsid group A or subgroup Bii.
Figure 3. Phylogenetic tree of CAstVs based on partial ORF 1b amino acid sequences. The tree was constructed using Mega 4 (Tamura et al., Citation2007) using the neighbour-joining method and 1000 bootstrap replicates (bootstrap values are shown on the tree). Data relating to the origin of the CAstVs, including the Genbank accession numbers, are shown in . ANV-1 was used to root the tree.
Conserved and variable capsid regions
Multiple alignment capsid protein comparisons of CAstVs representing groups A and B revealed very low levels of conserved sequence. The longest conserved sequences included two sequences of seven amino acids located at residues 173 to 179 and 715 to 721 in the 11672 capsid sequence. In contrast, capsid sequence alignments of CAstVs from the same groups identified more extensive regions of conserved sequence as well as more variable regions. When the CAstVs from the Bi and Bii subgroups were compared, it was evident that the N-terminal regions comprising the first 415 amino acid residues were substantially more conserved and contained less variable regions than the C-terminal regions (residues 416 to end). Pairwise comparisons showed that the N-terminal regions (residues 1 to 415) of the subgroups Bi and Bii displayed 90 to 92% amino acid identity, whereas the remaining C-terminal regions displayed 76 to 78% identity (). Similar comparisons involving the A subgroups also showed that the N-terminal regions were more conserved (80 to 89%) than the C-terminal regions (72 to 75%). Several variable regions were identified when the C-terminal regions of representative CAstVs from subgroups Bi and Bii were compared (). These included three short hypervariable stretches, denoted A, B and C in , where, in each case, the majority of amino acids were found to differ. Similar comparisons of CAstVs from Group A also demonstrated the presence of conserved and variable regions (alignment not shown), including those at residues 535 to 549 and 621 to 634 (as numbered in the 612 capsid sequence).
Figure 4. Clustal W alignment of C-terminal amino acid (residues 416 to end) regions of the capsid proteins of six representative group B CAstVs. The 11672, FP3 and VF06-7/5 belong to subgroup BI, and the UGA-2006, 05V150/152/154 and VF07-4/2 belong to subgroup Bii. Three examples of variable regions, corresponding to residues (4a) 434 to 442, (4b) 531 to 545 and (4c) 647 to 655 of the 11672 capsid protein, are shown. Asterisks denote amino acids shared by all six representative CAstVs, while full stops and colons denote amino acids with shared characteristics.

Table 3. Pairwise comparison of CAstV subgroups showing amino acid identities of N-terminal (residues 1 to 415) and C-terminal (residues 416 to end) regions (in parentheses).
Discussion
This paper describes the first comparative sequence analysis of the complete capsid proteins of CAstVs. Most of the 25 CAstVs compared were from broiler chickens that originated in the UK, but examples from other European countries, South Africa and the USA were also included. Our investigation revealed considerable capsid protein sequence diversity, and this has implications for both CAstV diagnosis and vaccination-based control strategies.
Although a limited comparison of six partial ORF 2 (capsid) sequences has recently been described (Pantin-Jackwood et al., Citation2011), most of the CAstV sequence data available to date relate to partial ORF 1b sequences (Pantin-Jackwood et al., Citation2006; Smyth et al., Citation2010). In the absence of virus-specific antisera for antigenic characterization, the detection and identification of CAstVs have depended on sequence similarity shared with the published ORF 1b sequences. In earlier work we showed that, based on partial ORF 1b sequences, CAstVs originating in the USA, Europe and South Africa could be assigned to one of two major polymerase groups (I and II), which shared ~76 to 79% nucleotide identity (Smyth et al., Citation2009). In the present paper, we have shown that CAstVs, which share comparatively high levels of ORF 1b nucleotide (>75%) and amino acid (>80%) identities, can differ substantially in their capsid protein sequences. Thus, CAstVs can be assigned to one of two major capsid groups, A and B, which share 38 to 40% amino acid identity. It is noteworthy that these two capsid-based groups do not correspond to the two polymerase-based groups, such that CAstVs—which possess ORF 1b sequences that are characteristic of polymerase group I—can possess capsid sequences that are representative of group A or subgroup Bii, while CAstVs with ORF 1b sequences that are characteristic of polymerase group II have capsid sequences typical of subgroup Bi.
The taxonomic relationships of the CAstV capsid groups to one another and to other avastroviruses remain to be resolved. Although the capsid protein sequence identity (38 to 40%) shared by the CAstV groups is similar to those shared by TAstV-1 with the taxonomically distinct TAstV-2 (36%) and with ANV (41%) species, the ORF 1b amino acid identity (>80%) shared by CAstVs belonging to both capsid groups is substantially greater than that (~57%; determined in this study from TAstV-1 and TAstV-2 genomic sequences, Y15936 and AF206663) shared by the two TAstV species. A recent taxonomic proposal—submitted for approval (http://talk.ictvonline.org/files/proposals/taxonomy_proposals_vertebrate1/m/vert01/2358.aspx) to the International Committee for the Taxonomy of Viruses (ICTV) by members of the Astroviridae Study Group during the preparation of the 9th ICTV Report—states that a classification based on genetic criteria is more appropriate than classifying the viruses into species within the genus Avastrovirus based only on host of origin. Based on phylogenetic analysis of complete ORF 2 sequences, a new classification was proposed, which establishes three new species within the genus—namely Avastrovirus GI.A, including TAstV-1; Avastrovirus GI.B, including ANV-1 and ANV-2; and Avastrovirus GII.A, including TAstV-2 and DAstV. According to this new proposal and based on the amino acid identities determined in the present investigation, the group A and group B CAstVs will need to be recognized as two additional avastrovirus species. We note that this recent proposal does not consider the genetic relatedness exhibited by the more conserved regions of the avian astrovirus genome, such as ORFs 1a and 1b, encoding the non-structural proteins, and the untranslated 5′ and 3′ regions. If such was the case, a single CAstV species would be recognized that might be subdivided into major capsid groups A and B.
The CAstV capsid protein sequence diversity is likely to be reflected in substantial antigenic diversity. On the basis of the very low amino acid identity (38 to 40%), we would expect the CAstV capsids belonging to groups A and B to be antigenically distinct. Earlier work in this laboratory suggests that this is the case. Thus, heterologous antisera, which were raised by separately infecting groups of SPF chickens with the 612 (group A) and 11672 (group B) isolates, exhibited low-level IIF cross-reactivities when reacted with virus-infected primary chick embryo liver cells (Todd et al., Citation2009a). It is likely that the low-level cross-reactivities we detected in that study can be attributed to virus-specific antibodies that react with the non-structural RNA polymerase proteins, which share relatively high levels (>80%) of amino acid identity, or with the non-structural protease proteins.
Capsid sequence comparisons of the CAstVs belonging to each of the two capsid groups suggest that each may comprise more than one serotype. Earlier studies with human astrovirus and TAstV-2 showed that isolates can share as high as 85% capsid protein identity and still be considered serotypically different (Tang et al., Citation2005). Sequence comparisons of the group A CAstV capsids identified three subgroups (i, ii and iii), which showed inter-group amino acid identities of <85%, and this was also the case when the capsids of the Bi and Bii subgroups were compared. Work with human astroviruses has shown that the N-terminal region (residues 1 to 415) of the capsid protein is located in the inside of the capsid, while the C-terminal region (residues 416 to end) is located on the outside of the capsid, thereby facilitating its interactions with the host cell receptors and the host's immune response (Krishna, Citation2005). Evidence from human astrovirus studies also suggests that the C-terminal regions contain neutralizing epitopes (Sanchez-Fauquier et al., Citation1994; Bass & Upadhyapuda, Citation1997). On this basis, sequence differences in the C-terminal regions of the CAstV capsids are likely to be comparatively more important in relation to antigenic variation. Detailed examination of the C-terminal capsid regions (residues ~416 to end) showed that the Bi and Bii subgroups shared 76 to 78% amino acid identity, while pairwise comparisons of the A subgroups i, ii and iii estimated values that were less than 76%. We suspect that the extents of these C-terminal sequence variations exhibited by the A and B subgroups are likely to be manifested as serotypic differences. However, serological evidence will be required to confirm that this is the case.
Because the majority of CAstVs characterized were detected in the UK since 2004, our investigation into capsid diversity is relatively limited in terms of geographical origin. The CAstVs detected in UK samples can be assigned to two B subgroups and three A subgroups. CAstVs belonging to the Bii subgroup were obtained from the USA (Pantin-Jackwood et al., Citation2011) and Germany (05V150/152/154), while the CAstVs from the Netherlands (P22.18.00) and South Africa (612 isolate) can be assigned to subgroup Ai. Detailed analysis of CAstVs obtained from pooled intestinal contents of longitudinal surveys has shown that viruses from both capsid groups (VF08-29 and VF08-54) and different A subgroups (VF08-54 and VF08-60) can be found in the same broiler flock. These findings are consistent with the common occurrence in broiler and parent flocks of antibodies to both CAstV groups (Todd et al., Citation2009b), and we consider it likely that infections with all five subgroups recognized to date and other, as yet undiscovered, subgroups will be found worldwide. Indeed, the variety of CAstVs within the survey pools may exceed that which has already been detected, since the pooling of samples and the likelihood of the primers amplifying the most abundant CAstVs will limit the number of different CAstVs detected. It should also be noted that although the adaptation of CAstVs to cell culture may involve the introduction of mutations, the CAstV isolates investigated in this paper are located in groups and subgroups together with field samples and are not noticeably displaced.
The levels of capsid sequence variation have implications for the diagnosis of CAstV infections and their control. Currently, we are using a real-time RT-PCR test, which quantitatively detects virus RNAs from all known CAstVs from capsid group A or B (Smyth et al., Citation2010). This test uses primers located in the highly conserved region close to the 3′ end of ORF 1b and encompassing the 24-nucleotide intergenic region. At present, RT-PCR tests that distinguish CAstVs from groups A and B have not been described. With regards to serology, IIF and virus neutralization tests have been described (Baxendale & Mebatsion, Citation2004; Todd et al., Citation2009b). Due to the low-level cross-reactivity exhibited by CAstVs from groups A and B, separate IIF tests are required to detect serum antibodies to all CAstV infections (Todd et al., Citation2009b). Although an antibody-detecting enzyme-linked immunosorbent assay (ELISA) based on cell-culture-grown virus has been described for ANV (Decaesstecker & Meulemans, 1991), to date a similar ELISA for CAstV has not been reported, mainly due to the relatively poor growth of CAstV in cell culture. However, preliminary work in our laboratory has shown that an ELISA based on a recombinant CAstV11672 capsid protein produced in insect cells by a recombinant baculovirus has the capability to detect antibodies induced by infection with CAstVs from subgroups Bi and Bii, but will not detect antibodies induced by infection with a Group A CAstV isolate (Todd, 2011 unpublished results).
Because CAstV infections have been implicated in growth retardation problems in broilers, including runting stunting syndrome, there is increasing interest in the development of CAstV vaccines. Vaccine development has been restricted due to the limited availability of cell-culture-adapted virus isolates and the absence of a suitable experimental disease virus challenge model with which to test vaccine efficacy. Attempting to overcome these problems, Sellers et al. (Citation2010) have recently shown that vaccination of breeders with a purified recombinant baculovirus-expressed CAstV capsid protein provided partial protection to runting stunting syndrome in progeny broiler chickens. Vaccinating breeders during rear should not only ensure that chicks are hatched with uniformly high levels of maternal antibody but should also limit CAstV infections of in-lay breeder chickens, thereby inhibiting vertical transmission of CAstV via the embryo to the hatched chick. The detection of CAstVs from subgroup Bi in dead-in-shell chicks (FP3 isolate; Spackman et al., Citation1984) and weak 1-day-old chicks (11672, 11522 and 1010 isolates) supports the view that these viruses are vertically transmitted and that they may contribute to hatchability problems. Based on the capsid sequence diversity, the antigenic variation exhibited by CAstVs is likely to be substantial and will need to be taken into consideration if vaccine development is being contemplated.
Acknowledgements
This work was funded in part by the Biotechnology and Biological Sciences Research Council, the Department of the Environment, Food and Rural Affairs, and the Department of Agriculture and Rural Development for Northern Ireland.
References
- Bass , D.M. and Upadhyayula , U. 1997 . Characterization of human serotype 1 astrovirus-neutralizing epitopes . Journal of Virology , 71 : 8666 – 8671 .
- Baxendale , W. and Mebatsion , T. 2004 . The isolation and characterisation of astroviruses from chickens . Avian Pathology , 33 : 364 – 370 .
- Decaesstecker , M. and Meulemans , G. 1991 . An ELISA for the detection of antibodies to avian nephritis virus and related entero-like viruses . Avian Pathology , 20 : 523 – 530 .
- Guy , J.S. , McNulty , M.S. and Hayhow , C.S. 2008 . “ Avian enterovirus-like viruses ” . In Diseases of Poultry , 12th edn , Edited by: Saif , Y.M. , Fadly , A.M. , Glisson , J.R. , McDougald , L.R. , Nolan , L.K. and Swayne , D.E. 356 – 361 . Ames : Iowa State Press .
- Imada , T. , Yamaguchi , S. , Mase , M. , Tsukamoto , K. , Kubo , M. and Morooka , A. 2000 . Avian nephritis virus (ANV) as a new member of the family Astroviridae and construction of infectious ANV cDNA . Journal of Virology , 74 : 8487 – 8493 .
- Kang , K.I. , Icard , A.H. , Linnemann E. , Sellers , H.S. and Mundt , E. , 2012 , 44 Determination of the full length sequence of a chicken astrovirus suggests a different replication mechanism . Virus Genes , 45 – 50
- Krishna , N. K. 2005 . Identification of structural domains involved in astrovirus capsid biology . Viral Immunology , 18 : 17 – 26 .
- Larkin , M.A. , Blackshields , G. , Brown , N.P. , Chenna , R. , McGettigan , P.A. , McWilliam , H. , Valentin , F. , Wallace , I.M. , Wilm , A. , Lopez , R. , Thompson , J.D. , Gibson , T.J. and Higgins , D.G. 2007 . Clustal W and Clustal X version 2.0 . Bioinformatics , 23 : 2947 – 2948 .
- McNeilly , F. , Connor , T.J. , Calvert , V.M. , Smyth , J.A. , Curran , W.L. , Morley , A.J. , Thompson , D. , Singh , S. , McFerran , J.B. , Adair , B.M. and McNulty , M.S. 1994 . Studies on a new enterovirus-like virus isolated from chickens . Avian Pathology , 23 : 313 – 327 .
- McNulty , M.S. , Connor , T.J. , McNeilly , F. and McFerran , J.B. 1991 . Biological characterisation of avian enteroviruses and enterovirus-like viruses . Avian Pathology , 19 : 75 – 87 .
- Pantin-Jackwood , M.J. , Spackman , E. and Woolcock , P.R. 2006 . Molecular characterization and typing of chicken and turkey astroviruses circulating in the United States: implications for diagnostics . Avian Diseases , 50 : 397 – 404 .
- Pantin-Jackwood , M.J. , Strother , K.O. , Mindt , E. , Zsak , L. , Day , J.M. and Spackman , E. 2011 . Molecular characterization of avian astroviruses . Archives of Virology , 156 : 235 – 244 .
- Sanchez-Fauquier , A. , Carrascosa , A.L. , Carrascosa , J.L. , Otero , A. , Glass , R.I. , Lopez , J.A. , San Martin , C. and Melero , J.A. 1994 . Characterization of a human astrovirus serotype 2 structural protein (VP26) that contains an epitope involved in virus neutralization . Virology , 201 : 312 – 320 .
- Sellers , H. , Linneman , E. , Icard , A. H. and Mundt , E. 2010 . A purified recombinant baculovirus expressed capsid protein of a new astrovirus provides partial protection to runting-stunting syndrome in chickens . Vaccine , 28 : 1253 – 1263 .
- Smyth , J.A. , Connor , T.J. , McNeilly , F. , Moffet , D.A. , Calvert , V.M. and McNulty , M.S. 2007 . Studies on the pathogenicity of enterovirus-like viruses in chickens . Avian Pathology , 36 : 119 – 126 .
- Smyth , V.J. , Jewhurst , H.L. , Adair , B.M. and Todd , D. 2009 . Detection of chicken astrovirus by reverse transcription polymerase chain reaction . Avian Pathology , 38 : 293 – 299 .
- Smyth , V.J. , Jewhurst , H.L. , Wilkinson , D.S. , Adair , B.M. , Gordon , A.W. and Todd , D. 2010 . Development and evaluation of real-time TaqMan® RT-PCR assays for the detection of avian nephritis virus and chicken astrovirus in chickens . Avian Pathology , 39 : 467 – 474 .
- Spackman , D. , Gough , R.E. , Collins , M.S. and Lanning , D. 1984 . Isolation of an enterovirus-like agent from the meconium of dead-in-shell chicken embryos . The Veterinary Record , 114 : 216 – 218 .
- Tamura , K. , Dudley , J , Nei , M. & Kumar , S. , 2007 MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0 . Molecular Biology and Evolution , 24 , 1596 – 1599 . (Publication PDF at http://www.kumarlab.net/publications)
- Tang , Y. , Murgia , M.V. and Saif , Y.M. 2005 . Molecular characterization of the capsid gene of two serotypes of turkey astrovirus . Avian Diseases , 49 : 514 – 519 .
- Todd , D. , Smyth , V.J. , Ball , N.W. , Donnelly , B.M. , Wylie , M. , Knowles , N.J. and Adair , B. 2009a . Characterisation of chicken enterovirus-like viruses, duck hepatitis virus (DHV) type 2 and DHV type 3 as astroviruses . Avian Pathology , 38 : 21 – 29 .
- Todd , D. , Wilkinson , D.S. , Jewhurst , H.L. , Wylie , M. , Gordon , A.W. and Adair , B.M. 2009b . A seroprevalence investigation of chicken astrovirus infections . Avian Pathology , 38 : 301 – 309 .
- Todd , D. , Trudgett , J. , McNeilly , F. , McBride , N. , Donnelly , B. , Smyth , V.J. , Jewhurst , H.J. and Adair , B.M. 2010 . Development and application of an RT-PCR test for detecting avian nephritis virus . Avian Pathology , 39 : 207 – 213 .
- Todd , D. , Trudgett , J.S. , Smyth , V.J. , Donnelly , B. , McBride , N. and Welsh , M.D. 2011 . Capsid protein sequence diversity of avian nephritis virus . Avian Pathology , 40 : 249 – 259 .