Abstract
Mycoplasma iowae is primarily a pathogen of turkeys and, although uncommon, it still persists in some areas of the world, where it may cause embryo mortality and leg lesions. A species-specific diagnostic polymerase chain reaction was developed using a forward primer based in the intergenic spacer region between the 16S rRNA and the 23S rRNA ribosomal genes and a reverse primer located within the 23S rRNA gene. The polymerase chain reaction proved to be both sensitive and specific. It detected M. iowae DNA in the six reference strains of serotypes I, J, K, N, Q and R and in 28 field isolates. With the six serotypes the test detected between 1 and 5 pg of M. iowae DNA. There were no non-specific reactions with the other avian Mycoplasma species. When the closest phylogenetically related species were checked, a weak reaction with Mycoplasma muris was observed that disappeared when the annealing temperature was increased by 2°C.
Introduction
Mycoplasma iowae has been isolated from turkeys and chickens (Bradbury & Kleven, Citation2008) and has been associated with embryo mortality and leg lesions in experimentally infected turkeys and chickens (Bradbury & Ideris, Citation1982; Bradbury & McCarthy, Citation1984; Bradbury et al., Citation1988). In the poultry industry, M. iowae is primarily a pathogen of turkeys causing reduced hatchability and late embryo mortality (Grant, Citation1987), resulting in a reduction in hatchability of 2 to 5% (Baxter-Jones, Citation1991). Natural infection is rarely seen in chickens (Al-Ankari & Bradbury, Citation1996). M. iowae infection used to be common in turkey flocks in Europe and North America and other parts of the world (Al-Ankari & Bradbury, Citation1996) but its occurrence was much reduced by industry eradication schemes, although it is still found occasionally in turkeys.
In 1994 Trampel and Goll reported an outbreak of M. iowae infection in 17-day-old male commercial turkey poults in the USA in which leg weakness and dehydration were seen, together with chondrodystrophy of the hock joints, accumulation of fluid in the hock joint, valgus deformities, shortening and bowing of the tarsometatarsal bones and curled toes (Trampel & Goll, Citation1994). Dunn et al. (Citation2005) observed several cases of uneven growth and leg problems in commercial turkeys of ages 3 days to 3 weeks with leg lesions suggestive of chondrodystrophy. M. iowae was identified in swabs from the joints. Also in the USA, Ley et al. (Citation2010) described an association between M. iowae and a range of skeletal lesions including vertebral lesions and chondrodystrophy in commercial turkeys. They suggested that M. iowae should be considered in differential diagnosis in such cases.
Efficient diagnostic tests are therefore necessary for detecting infection and establishing and maintaining M. iowae-free stock but turkeys and chickens have a poor antibody response to M. iowae (Bradbury & McCarthy, Citation1984; Bradbury et al., Citation1988) and serological screening has not proved useful for screening flocks (Kiss et al., Citation1997), possibly because of the antigenic diversity amongst M. iowae strains (Rhoades, Citation1984). Traditionally diagnosis has relied on isolation and identification of the microorganism, although this can be a time-consuming procedure (García et al., Citation1997). Furthermore, the antigenic diversity within the species can complicate the identification procedure (Al-Ankari & Bradbury, Citation1996), because M. iowae encompasses six serotypes: I, J, K, N, Q, and R (Dierks et al. Citation1967).
In view of these diagnostic problems, polymerase chain reactions (PCRs) for detecting M. iowae have been described several of which are based on the 16S rRNA gene. Both conventional (Zhao & Yamamoto, Citation1993; Kempf et al., Citation1994; Boyle et al., Citation1995; Lierz et al., Citation2008) and real-time (Cai et al., Citation2008) M. iowae-specific PCRs have been described. Others have used non-specific broad-range primers and have required further analysis such as restriction fragment length polymorphism (Fan et al., Citation1995; García et al., 1996; Kiss et al., Citation1997). Laigret et al. (Citation1996) designed a specific M. iowae PCR based on a sequence upstream of the rRNA gene operon, and Raviv & Kleven (Citation2009) used the same region to design a real-time PCR.
In earlier unrelated studies the intergenic spacer region (ISR) located between the 16S and 23S rRNA genes has been sequenced for most members of the genus Mycoplasma (Harasawa et al., Citation2004; Volokhov et al., Citation2006; Ramírez et al., Citation2008) and, as was expected due to the fewer evolutionary constraints on the region (Barry et al., Citation1991), there was greater inter-species variation in the ISR than in the 16S RNA gene, making the former a potentially better target for species-specific PCRs than the latter.
Thus the ISR has been used successfully as the target for a variety of species-specific mycoplasma PCRs; for example Mycoplasma pulmonis in rat clinical isolates (Takahashi-Omoe et al., Citation2004), Mycoplasma felis directly in feline clinical samples (Chalker et al., Citation2004) and Mycoplasma synoviae from culture or swabs (Ramírez et al., Citation2006). Furthermore, in a previous study, the ISR sequences of 22 M. iowae strains including all six serotypes were determined and all showed complete similarity to the published sequence AJ780997 (Ramírez et al., Citation2008). The highest similarity within the ISRs of the avian mycoplasmas with respect to M. iowae was 47.8% with Mycoplasma gypis. When the ISR sequence of M. iowae was compared with those of mammalian Mycoplasma species, Mycoplasma microti, Mycoplasma penetrans and Mycoplasma muris, which are the species most closely related by 16S rDNA-based phylogeny (Johansson & Pettersson, Citation2002), the similarities were 79.3%, 65.5% and 62.3% respectively.
This paper describes a species-specific PCR that was then developed for M. iowae with the forward primer based in the ISR and the reverse primer within the 23S rRNA gene using the same strategy that was employed to develop a specific PCR for M. synoviae (Ramírez et al., Citation2006). Its efficacy was confirmed with 34 M. iowae strains including the six reference serotypes and with 23 other avian Mycoplasma species, plus M. penetrans and M. muris.
Materials and Methods
Organisms and culture conditions
The type strains of 23 avian Mycoplasma species used were the following: Mycoplasma anatis 1340T, Mycoplasma anseris 1219 T, Mycoplasma buteonis BbT2gT, Mycoplasma cloacale 383T, Mycoplasma columbinasale 694T, Mycoplasma columbinum MMP-1T, Mycoplasma columborale MMP-4T, Mycoplasma corogypsi Bv1T, Mycoplasma falconis H/T1T, Mycoplasma gallinaceum DDT, Mycoplasma gallinarum PG16T, Mycoplasma gallisepticum PG31T, Mycoplasma gallopavonis WR1T, Mycoplasma glycophilum 486T, M. gypis B1/T1T, Mycoplasma imitans 4229T, Mycoplasma iners PG30T, M. iowae 695T, Mycoplasma lipofaciens R171T, Mycoplasma meleagridis 17529T, Mycoplasma pullorum CKKT, Mycoplasma sturni UCMFT and M. synoviae WVU 1853T. presents information for the six M. iowae serotypes (including the type strain) and 28 M. iowae field strains isolates from turkeys and chickens, isolated over a range of years and from different geographical locations.
Table 1. M. iowae type strain, reference strains and field strains.
A mycoplasma isolated from penguins, provisionally named “Mycoplasma sphenisci” with a type strain designated as UCMJ (Frasca et al., Citation2005) was also included together with two mammalian Mycoplasma species—M. penetrans GTU54T and M. muris RIII4T—that are phylogenetically related to M. iowae.
Mycoplasmas were cultivated as previously described by Bradbury (Citation1977) and the identity of all the strains was confirmed by an indirect fluorescent antibody test (Rosendal & Black, Citation1972) after cloning by filtration.
DNA extraction and measurement
DNA was extracted from 1.5 ml of each mycoplasma culture, after centrifuging and washing, using Chelex resin as described in a previous paper (Ramírez et al., Citation2006), and the DNA of the different M. iowae strains was quantified with a fluorometer (DyNA Quant™ 200; Hoefer, Pharmacia Biotech Inc. Piscataway, NJ, USA) following the manufacturer's instructions.
Primer design and use in amplification of the 16S to 23S ISR
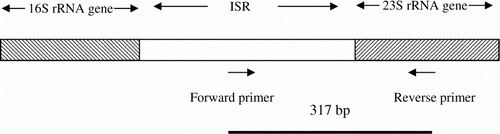
M. iowae forward primer (MiF) targeting the ISR was designed manually based on the avian mycoplasma ISR sequences (Genbank accession numbers AB98503, AB098504 and AJ780982 to AJ781002). The reverse primer was also chosen manually from the 5′ end of the 23S rRNA gene when the sequences of this region were compared (unpublished data). From these sequences, two M. iowae species-specific regions were identified. Gene Runner (version 3.05; Hastings Software, Inc. http://www.generunner.net/ accessed on 25/05/12) was used to ensure that there was no cross-reactivity between MiF or M. iowae reverse primer (MiR) primers with the ISRs and the 23S rRNA genes of avian Mycoplasma species other than M. iowae, The same was done with the phylogenetically related mycoplasmas mentioned above, using the sequences of the ISR for which accession numbers were D89508 (M. penetrans), D89507 (M. muris), FJ609188 (M. microti) and the sequence of the 23S rRNA gene of M. penetrans (EF061774). The sequences of the selected forward and reverse primers were: MiF, 5′-TAA TTG TAA TAA CAG GTC GGA TTC-3′; and MiR, 5′-CTA GGT ATA GCT TCA TTA ACC-3′. In addition, a BLAST search was carried out with both primers to prevent other non-specific reactions. The expected PCR product size was 317 base pairs and its location is illustrated in .
DNA amplifications were performed in a DNA thermal cycler (Perkin-Elmer Applied Biosystems, PE Biosystems, Foster City, CA, USA). Reaction mixtures contained 2.5 U Taq DNA polymerase (ABgene, Fisher Scientific UK Ltd, Loughborough, Leicestershire, UK), 0.2 µM each primer, 1×reaction buffer, 1.75 mM MgCl2, 0.2 mM dNTPs, and water to a volume of 50 µl. One microlitre of the template was added. DNA amplification was achieved with five cycles of denaturation at 94°C for 15 sec, renaturation at 60°C for 30 sec, and elongation at 72°C for 2 min, followed by 30 cycles the same as before but with an extension of 2 sec per cycle in the elongation step. A 5 µl amount of each amplified product was separated by electrophoresis on a 1.5% agarose gel. Gels were stained with ethidium bromide (3 µg/ml) and visualized with ultraviolet light.
Sensitivity and specificity of the M. iowae assay
The sensitivity was tested with DNA-extracted broth cultures of all six M. iowae serotypes using 10-fold dilutions up to 10−9. The DNA was measured as described above, but only in the most concentrated dilution; the amounts of DNA in the remaining dilutions were then deduced and PCRs were carried out on each dilution. Viable counts were carried out in parallel for each culture (Bradbury, Citation1977) and the numbers of colony-forming units (CFUs) per 100 µl were calculated. The PCR was then applied to 28 M. iowae field isolates.
The specificity of the PCR was verified using cultures of the 22 other recognized avian Mycoplasma species plus “M. sphenisci” and the two phylogenetically related mammalian mycoplasmas, M. muris and M. penetrans.
Results
Testing the primer sets
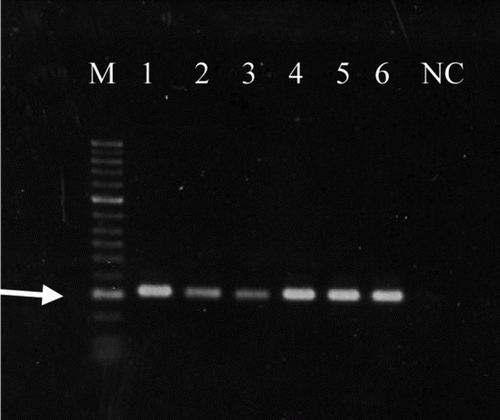
The BLAST analysis of the primers did not reveal significant similarity to any other sequence in the GenBank. shows that the MiF-23S PCR produced the expected fragment (317 base pairs) () from all six M. iowae serotypes and also from the 28 field isolates (data not shown).
Figure 2. M. iowae PCR result with the six M. iowae serotypes. M, marker (arrow indicates 300 base pairs); lane 1, M. iowae 695 (serotype I); lane 2, M. iowae DJA (serotype J); lane 3, M. iowae DK-CPA (serotype K); lane 4, M. iowae FMN (serotype N); lane 5, M. iowae L3-10B (serotype Q); lane 6, M. iowae DRA-0 (serotype R); NC, negative control.
Sensitivity and specificity of the M. iowae assay
The specific M. iowae PCR was clearly positive with strain 695T up to and including the 10−4 dilution (), which corresponds with a mean of nearly 5 CFUs per PCR reaction, and with an approximate DNA amount of 11 pg (). At the 10−5 dilution there was a very faint band that cannot be seen in , thus detecting 1 pg DNA and with less than 1 CFU per reaction. The reference strain of serotype J produced clear positive results up to the 10−2 (30 pg) dilution and a weak band at 10−3 (3 pg). Reference strains of serotypes K, N, Q and R were all clearly positive at the 10−3 dilution (mean: 47 pg) and faintly positive at the 10−4 dilutions (mean: 4.7 pg).
Figure 3. Sensitivity of the M. iowae PCR tested on eight dilutions of M. iowae 695T (serotype I). M, marker (arrow indicates 300 base pairs); lane 1, undiluted; lane 2, 10−1; lane 3, 10−2 lane 4, 10−3; lane 5, 10−4; lane 6, 10−5; lane 7, 10−6 lane 8, 10−7; NC, negative control; PC, positive control.

Table 2. Viable counts and amount of DNA used for testing sensitivity of the M. iowae PCR assay with M. iowae serotypes
The M. iowae PCR was highly specific, reacting only with M. iowae template and with no amplified PCR products from the other avian mycoplasma species tested (data not shown).
No false positive reactions occurred with M. penetrans or with “M. sphenisci” but there was a faint reaction with M. muris. The product was slightly larger than the expected size and disappeared if the annealing temperature was increased to 62°C, whereas the 685 strain remained positive.
Discussion
Several of the PCR assays described for M. iowae have been based on the 16S rRNA gene (Kempf et al., Citation1994; Boyle et al., Citation1995; Kiss et al., Citation1997; Cai et al., Citation2008; Lierz et al., Citation2008) although two (Laigret et al., Citation1996; Raviv & Kleven, Citation2009) targeted a region upstream of the rRNA operon. The forward primer of the PCR assay described in this paper is based on the ISR between the 16S and 23S rRNA genes and the reverse primer on the 5′ end of the 23S rRNA gene. Takahashi-Omoe et al. (Citation2004) found that when a specific primer pair with one primer targeting the ISR was used, the diagnostic accuracy improved over PCRs based in the 16S rRNA gene because it reduced the possibility of cross-reactions, particularly between closely-related species such as M. gallisepticum and M. imitans whose 16S rDNA sequences are nearly identical.
Another advantage of ISR-based PCRs is that this region has been the focus of attention in mycoplasmology and the sequences of most Mycoplasma species have been deposited, whereas little is known about the region upstream of the rRNA operons. An interesting feature was the lack of variability found in the M. iowae ISR sequences of all of 22 strains tested, including the six serotypes (Ramírez et al., Citation2008). Earlier the ISR was found useful in differentiating between two biovars of Ureaplasma urealyticum while the serovars were found to be identical (Harasawa & Kanamoto, Citation1999; Kong et al., Citation1999). Based on this and other data, Robertson et al. (Citation2002) published the division of the biovars into two separate species, U. urealyticum and Ureaplasma parvum.
The M. iowae PCR assay described here is both sensitive and specific when examining cultured mycoplasmas. Using the reference strains, it could detect between 1 and 5 pg of M. iowae DNA, which approximately corresponded to between 0.2 and 2.99 CFUs per µl (). The complete genome sequence of M. iowae has been published recently (Wei et al., Citation2012) and reveals the presence of two rRNA operons, which may help to explain why the sensitivity can be less than one organism per reaction. This detection limit is similar to that obtained by Lierz et al. (Citation2008) who detected as little as 0.1 CFU, and is lower than the 3.2 CFUs per PCR reaction found by Cai et al. (Citation2008) or the 10 colour changing units per reaction reported by Laigret et al. (Citation1996). Zhao & Yamamoto (Citation1993) could detect 1 pg of M. iowae DNA, while Boyle et al. (Citation1995) and Kempf et al. (Citation1994) did not assess the amount of DNA or count the CFUs. Raviv & Kleven (Citation2009) reported a sensitivity of 10 copies for their real-time M. iowae PCR. They found that the ISR was not suitable for real-time TaqMan PCR because of its high A/T content and difficulties in reaching the required melting temperatures of the oligonucleotides. The specificity of our PCR test was demonstrated, in that there were no reactions with any of 22 other recognized avian Mycoplasma species and “M. sphenisci”. These included the two avian species (M. gallisepticum and M. imitans) that are in the same phylogenetic “pneumoniae group” as M. iowae based on the 16S rDNA gene sequences (Brown et al., Citation2011) although they are not in the same cluster. Two other non-avian mycoplasmas, M. penetrans and M. muris, were analysed because they are more closely related, being in the same phylogenetic cluster as M. iowae (Brown et al., Citation2011), and a weak reaction resulted with the closest known relative, M. muris (a mycoplasma isolated from rodents). The PCR product was slightly larger than the expected size and was possibly due to amplification of a different part of the genome since the in silico tests had shown no match within the ISR. If the annealing temperature was increased by 2°C the non-specific reaction disappeared, but this measure should not be necessary when investigating samples of avian origin. There was no reactivity with M. penetrans, and M. microti was not evaluated. Of all the other M. iowae PCRs mentioned above, Laigret et al. (Citation1996) were the only ones to check and rule out reactivity with M. muris. Mycoplasma PCRs based on the 16S rDNA may present particular difficulties with species-specific reactivity. For example, Kempf et al. (Citation1994) reported non-specific amplification of M. gallisepticum DNA with their 16S rDNA-based M. iowae primers. Zhao & Yamamoto also (1993) reported difficulties in establishing the perfect PCR conditions for a specific reaction although their target DNA was not defined.
The PCR reported here is both sensitive and specific for M. iowae and it should have sufficient sensitivity for clinical use. M. iowae can be present in embryos and sometimes in the lesions of live birds. It is also found in the gastrointestinal tract, cloaca and other samples (Bradbury & Kleven, Citation2008). Further work applying this PCR assay to suitable field samples should be carried out but in Europe this is difficult due to the rarity of the infection. Nevertheless, this assay has the potential to provide a further useful diagnostic tool for the detection of M. iowae.
References
- Al-Ankari , A.S. and Bradbury , J.M. 1996 . Mycoplasma iowae: a review . Avian Pathology , 25 : 205 – 229 .
- Barry , T. , Colleran , G. , Glennon , M. , Dunican , L.K. and Gannon , F. 1991 . The 16S/23S ribosomal spacer region as a target for DNA probes to identify eubacteria . Genome Research , 1 : 51 – 56 .
- Baxter-Jones , C. 1991 . “ Egg hygiene: microbial contamination, significance and control ” . In Avian Incubation , Edited by: Tullet. , S.G. Vol. 22 , 269 – 276 . London : Poultry Science Symposium Series, Butterworth-Heinemann .
- Boyle , J.S. , Good , R.T. and Morrow , C.J. 1995 . Detection of the turkey pathogens Mycoplasma meleagridis and M. iowae by amplification of genes coding for rRNA . Journal of Clinical Microbiology , 33 : 1335 – 13 .
- Bradbury , J.M. 1977 . Rapid biochemical tests for characterisation of the Mycoplasmatales . Journal of Clinical Microbiology , 5 : 531 – 534 .
- Bradbury , J.M. and Ideris , A. 1982 . Abnormalities in turkey poults following infection with Mycoplasma iowae . The Veterinary Record , 110 : 559 – 60 .
- Bradbury , J.M. and McCarthy , J.D. 1984 . Mycoplasma iowae infection in chicks . Avian Pathology , 13 : 529 – 543 .
- Bradbury , J.M. & Kleven , S.H. 2008 . Mycoplasma iowae infection . In Y.M. Saif , A.M. Fadly , J.R. Glisson , L.R. McDougald , L.K. Nolan & D.E. Swayne , Diseases of Poultry , 12th edn (pp. 231 – 236 ). Ames , , Iowa : Blackwell Publishing Professional .
- Bradbury , J.M. , Ideris , A. and Oo , T.T. 1988 . Mycoplasma iowae infection in young turkeys . Avian Pathology , 17 : 149 – 171 .
- Brown , D.R. , May , M. , Bradbury , J.M. , Balish , M.F. , Calcutt , M.J. , Glass , J.I. , Tasker , S. , Messick , J.B. , Johansson , K-E. and Neimark , H. 2011 . “ Genus I. Mycoplasma ” . In Bergey's Manual of Systematic Bacteriology , 2nd edn. , Edited by: Krieg , N.R. , Staley , J.T. , Brown , D.R. , Hedlund , B.P. , Paster , B.J. , Ward , N.L. , Ludwig , W. and Whitman , W.B. Vol. 4 , 575 – 613 . New York : Springer .
- Cai , H.Y. , Bell-Rogers , P. , Parker , L. , Ferencz , A. and Pozder , P. 2008 . Development and field validation of a Mycoplasma iowae real-time polymerase chain reaction assay . Journal of Veterinary Diagnostic Investigation , 20 : 230 – 235 .
- Chalker , V.J. , Owen , W.M.A. , Paterson , C.J.I. and Brownlie , J. 2004 . Development of a polymerase chain reaction for the detection of Mycoplasma felis in domestic cats . Veterinary Microbiology , 100 : 77 – 82 .
- Dierks , R.E. , Newman , J.A. and Pomeroy , B.S. 1967 . Characterization of avian mycoplasma . Annals of the New York Academy of Sciences , 143 : 170 – 189 .
- Dunn , P. , Wallner-Pendleton , E. & Love , B. 2005 . Case report: Mycoplasma iowae infection in commercial turkeys . Paper presented at the 77th Northeastern Conference on Avian Diseases , Ithaca , NY ( cited by Ley et al. 2010 ).
- Fan , H.H. , Kleven , S.H. and Jackwood , M.W. 1995 . Studies of intraspecies heterogeneity of Mycoplasma synoviae, M. meleagridis, and M. iowae with arbitrarily primed polymerase chain reaction . Avian Diseases , 39 : 766 – 777 .
- Frasca , S. Jr. , Scott Weber , E. , Urquhart , H. , Liao , X. , Gladd , M. , Cecchini , K. , Hudson , P. , May , M. , Gast , R.J. , Gorton , T.S. and Geary , S.J. 2005 . Isolation and characterization of Mycoplasma sphenisci sp. nov. from the choana of an aquarium-reared Jackass Penguin (Spheniscus demersus) . Journal of Clinical Microbiology , 43 : 2976 – 2979 .
- Garcia , M. , Jackwood , M.J. , Head , M. , Levisohn , S. and Kleven , S.H. 1996 . Use of species-specific oligonucleotide probes to detect Mycoplasma gallisepticum, M. synoviae, and M. iowae PCR amplification products . Journal of Veterinary Diagnostic Investigation , 8 : 56 – 63 .
- García , M. , Gerchman , I. , Meir , R. , Jackwood , M.W. , Kleven , S.H. and Levisohn , S. 1997 . Detection of Mycoplasma meleagridis and M. iowae from dead-in-shell turkey embryos by polymerase chain reaction and culture . Avian Pathology , 26 : 765 – 778 .
- Grant , M. 1987 . Significance, epidemiology and control methods of Mycoplasma iowae in turkeys . PhD Thesis, Council for National Academic Awards .
- Harasawa , R. and Kanamoto , Y. 1999 . Differentiation of two biovars of Ureaplasma urealyticum based on the 16S-23S rRNA intergenic spacer region . Journal of Clinical Microbiology , 37 : 4135 – 4138 .
- Harasawa , R. , Pitcher , D.G. , Ramírez , A.S. and Bradbury , J.M. 2004 . A putative transposase gene assigned in the 16S-23S rRNA intergenic spacer region of Mycoplasma imitans . Microbiology , 150 : 1023 – 1029 .
- Johansson , K.E. and Pettersson , B. 2002 . “ Taxonomy of Mollicutes ” . In Molecular Biology and Pathogenicity of Mycoplasmas , Edited by: Razin , S. and Herrmann , R. 1 – 29 . New York : Kluwer Academic/Plenum Publishers .
- Kempf , I. , Blanchard , A. , Guittet , M. and Bennejean , G. 1994 . Comparision of antigenic and pathogenic properties of Mycoplasma iowae strains and development of a PCR-based detection assay . Research in Veterinary Science , 56 : 179 – 185 .
- Kiss , I. , Matiz , K. , Kaszanyitzky , E. , Chavez , Y. and Johansson , K.E. 1997 . Detection and identification of avian mycoplasmas by polymerase chain reaction and restriction fragment length polymorphism assay . Veterinary Microbiology , 58 : 23 – 30 .
- Kong , F. , James , G. , Zhenfang , M. , Gordon , S. , Wang , B. and Gilbert , G. 1999 . Phylogenetic analysis of Ureaplasma urealyticum – support for the establishment of a new species, Ureaplasma parvum . International Journal of Systematic and Evolutionary Microbiology , 49 : 1879 – 1889 .
- Laigret , F. , Deaville , J. , Bové , J.M. and Bradbury , J.M. 1996 . Specific detection of Mycoplasma iowae using polymerase chain reaction . Molecular and Cellular Probes , 10 : 23 – 29 .
- Ley , D. H. , Marusak , R.A. , Vivas , E.J. , Barnes , H.J. and Fletcher , O.J. 2010 . Mycoplasma iowae associated with chondrodystrophy in commercial turkeys . Avian Pathology , 39 : 87 – 93 .
- Lierz , M. , Hagen , N. , Lueschow , D. and Hafez , H.M. 2008 . Use of polymerase chain reactions to detect Mycoplasma gallisepticum, Mycoplasma imitans, Mycoplasma iowae, Mycoplasma meleagridis and Mycoplasma synoviae in birds of prey . Avian Pathology , 37 : 471 – 476 .
- Ramírez , A.S. , Naylor , C.J. , Hammond , P.P. and Bradbury , J.M. 2006 . Development and evaluation of a diagnostic PCR for Mycoplasma synoviae using primers located in the intergenic spacer region and the 23S rRNA gene . Veterinary Microbiology , 118 : 76 – 82 .
- Ramírez , A.S. , Naylor , C.J. , Pitcher , D.G. and Bradbury , J.M. 2008 . High inter-species and low intra-species variation in 16S-23S rDNA spacer sequences of pathogenic avian mycoplasmas offers potential use as a diagnostic tool . Veterinary Microbiology , 128 : 279 – 287 .
- Raviv , Z. and Kleven , S.H. 2009 . The development of diagnostic real-time TaqMan PCRs for the four pathogenic avian mycoplasmas . Avian Diseases , 53 : 103 – 107 .
- Rhoades , K. R. 1984 . Comparison of strains of Mycoplasma iowae . Avian Diseases , 28 : 710 – 717 .
- Robertson , J.A. , Stemke , G.W. , Davis , Jr J.W. , Harasawa , R. , Thirkell , D. , Kong , F. , Shepard , M.C. and Ford , D.K. 2002 . Proposal of Ureaplasma parvum sp. nov. and emended description of Ureaplasma urealyticum (Shepard et al. 1974) Robertson et al. 2001 . International Journal of Systematic and Evolutionary Microbiology , 52 : 587 – 597 .
- Rosendal , S. and Black , F. T. 1972 . Direct and indirect immunofluorescence of unfixed and fixed mycoplasma colonies . Acta Pathologica, Microbiologica et Immunologica Scandinavica , B80 : 615 – 622 .
- Takahashi-Omoe , H. , Omoe , K. , Matsushita , S. , Kobayashi , H. and Yamamoto , K. 2004 . Polymerase chain reaction with a primer pair in the 16S-23S rRNA spacer region for detection of Mycoplasma pulmonis in clinical isolates . Comparative Immunology, Microbiology and Infectious Diseases , 27 : 117 – 128 .
- Trampel , D.W. and Goll , F. Jr. 1994 . Outbreak of Mycoplasma iowae infection in commercial turkey poults . Avian Diseases , 38 : 905 – 909 .
- Volokov , D.V. , George , J. , Liu , S.X. , Ikonomi , P. , Anderson , C. and Chizhikov , V. 2006 . Sequencing of the intergenic 16S-23S rRNA spacer (ITS) region of Mollicutes species and their identification using microarray-based assay and DNA sequencing . Applied Microbiology and Biotechnology , 71 : 680 – 698 .
- Wei , S. , Guo , Z. , Li , T. , Zhang , T. , Li , X. , Zhou , Z. , Li , Z. , Liu , M. , Luo , R. , Bi , D. , Chen , H. , Zhou , R. and Jin , H. 2012 . Genome sequence of Mycoplasma iowae strain 695, an unusual pathogen causing deaths in turkeys . Journal of Bacteriology , 194 : 547 – 548 .
- Zhao , S. and Yamamoto , R. 1993 . Amplification of Mycoplasma iowae using polymerase chain reaction . Avian Diseases , 37 : 212 – 217 .