Abstract
Newcastle disease (ND) is an endemic disease in rural poultry of Western Africa. It may cause severe economic losses in the poultry sector and, as such, is listed as a notifiable disease by the World Organisation for Animal Health (OIE). Recently, a new genetic lineage of ND viruses was discovered in Western Africa. We determined the complete fusion (F) gene coding sequence of 12 ND viruses isolated from pigeons and rural chickens in six Nigerian states in 2007 and 2008. Phylogenetic analysis of the complete F coding sequence confirmed the circulation of genetically diverse ND isolates in a large geographic area in Nigeria. Next to isolates belonging to lineage 4b, viruses of the recently discovered lineage 7 (some of which were previously reported to escape routine real-time reverse transcriptase-polymerase chain reaction detection) were isolated in six states during the two-year period. The documented genetic variants occurred over a large geographic area, indicating an endemic circulation of these viruses. Three different velogenic fusion gene cleavage site motifs were observed. These findings confirm the endemic circulation and diversification of ND isolates in rural poultry and pigeons in Nigeria and highlight the importance of surveillance in developing countries to monitor the validity of rapid molecular diagnostic tools and of vaccination regimes.
Introduction
Newcastle disease (ND) is caused by virulent avian paramyxovirus type 1 (APMV-1) strains (Paramyxoviridae, Avulavirus) (Lamb et al., Citation2005). It is one of the most important notifiable poultry diseases listed by the World Organisation for Animal Health (OIE), with the potential to cause severe economic losses in the poultry sector worldwide, and affecting many species of birds. The clinical signs and severity of disease that different strains cause in birds vary widely. According to their virulence in poultry, ND isolates can be classified as lentogenic, mesogenic or velogenic (Alexander, Citation2003). Velogenic strains cause severe disease and high mortality in poultry. The pathogenicity in poultry can be determined using in vivo tests, such as the intracerebral pathogenicity index. Alternatively, the determination of the amino acid sequence at the cleaving site of the fusion gene precursor glycoprotein F0 is recognized for the determination of pathogenicity (OIE, Citation2008). The presence of multiple basic sequences (arginine R or lysine K) located at the C terminus of the F1 protein and phenylalanine (F) at the N terminus of the F2 protein is indicative of high pathogenicity (Alexander, Citation2000).
Similar to other single-stranded RNA viruses, avian paramyxoviruses have a significant genetic variability. Recently, two major genetic APMV-1 lineages have been described (Czegledi et al., Citation2006). Class I viruses of low virulence have been isolated from live bird markets and wild waterfowl, while class II constitutes the vast majority of viruses isolated from wild birds and poultry, including viruses of high virulence (Czegledi et al., Citation2006; Kim et al., 2007). Another study (Aldous et al., Citation2003) classifies APMV-1 into six distinct lineages (1 to 6). The majority of ND virus isolates responsible for outbreaks in Europe, Asia, and Southern Africa were classified as lineages 3 to 5. ND was historically reported to be widespread (Lancaster & Alexander, Citation1975) and continues to be the most important disease in commercial and rural chickens in West Africa (Bebay, Citation2006). In Nigeria, larger poultry farms tend to follow vaccination programmes against APMV-1, while small rural and backyard farms throughout the country normally do not (Adene & Oguntade, Citation2006). Poultry farming is an important industry in Nigeria, and is essential for subsistence of many small producers (Adene & Oguntade, Citation2006). To a lesser extent, pigeon farming for meat contributes to the subsistence of small producers (Adene & Oguntade, Citation2006).
Although ND has been officially and unofficially reported in the whole of the African continent, until recently virological and epidemiological data from Central Africa and Western Africa remained scarce. Two recent surveys covered the latter region, including samples from rural chickens and live bird markets in the period 2006 to 2008 (Snoeck et al., Citation2009; Cattoli et al., Citation2010). In both studies, ND viruses were isolated and genetically characterized from multiple countries in Western Africa and Central Africa, including Nigeria; and a genetically divergent new lineage, tentatively called lineage 7, was observed that seems to be restricted to Western Africa and to circulate in rural chickens and live bird markets during the period of investigation (Cattoli et al., Citation2010). Snoeck et al. (Citation2009) also documented lentogenic viruses of lineages 1, 2, and 3 that are genetically almost similar to vaccine strains, and additionally a single lineage 4b isolate in pigeons in south-western Nigeria. The aim of the present study was the detailed molecular characterization of recently circulating ND virus isolates in Nigerian rural poultry and pigeons, with a focus on the poorly studied viruses from the central and north-eastern regions of the country.
Materials and Methods
Viruses
Fifteen Nigerian virus isolates were selected from the virus repository of the National Veterinary Research Institute (Vom, Nigeria), and sent to the BSL3 laboratory of the Veterinary and Agrochemical Research Centre (VAR, Brussels, Belgium). They originated from 2007 to 2008 and covered a large geographic area (). The original samples were taken from pigeon and chicken (free-range local, live bird markets, and small-scale farms (<1000 birds, i.e. the non-vaccinated part of the Nigerian poultry sector). The 15 selected samples were found to be APMV-1-positive using virus isolation and haemagglutination inhibition testing at the National Veterinary Research Institute according to standard procedures (OIE, Citation2008). Upon arrival in the BSL3 laboratory of the VAR, the viruses were propagated in the allantoic cavities of 9-day-old to 11-day-old embryonated chicken eggs according to standard procedures (OIE, Citation2008) and were stored at −80°C.
Table 1. Identification, epidemiological data, and genotype of the ND virus isolates characterized in this study.
RNA extraction and reverse transcriptase-polymerase chain reaction
RNA was extracted from egg allantoic fluids using the High Pure Viral Nucleic acid kit (Roche, Vilvoorde, Belgium) according to the manufacturer's instructions, and RNA was stored at −80°C until use. The presence of APMV-1 RNA was confirmed using real-time reverse transcriptase-polymerase chain reaction (RT-PCR) as previously described (Kim et al., Citation2008). cDNA was produced from the viral RNA using the Transcriptor First Strand cDNA Synthesis Kit (Roche) in a 20 µl reaction volume containing 4 µl viral RNA, 60 µM final concentration of random hexamer primer, 1 mM each dNTP, and 10 u RT. The reaction mixture was incubated for 10 min at 25°C followed by 60 min at 50°C. Overlapping PCR products were produced using F-gene specific primers () and the Expand High Fidelity PCR System (Roche, Vilvoorde, Belgium). Then 1.5 mM MgCl2, 2.7 u DNA polymerase, 200 µM each dNTP, 500 nM F and R primers, and 1 µl cDNA were included in a final reaction volume of 50 µl. PCR amplification was performed using the following temperature protocol: 94°C for 2 min; 45 cycles of 94°C for 1 min, 45°C for 1 min and 68°C for 4 min; and a final extension at 72°C for 10 min.
Table 2. Oligonucleotides used for the PCR amplification of the F gene in overlapping fragments.
Sequencing and phylogenetic analysis
PCR products were visualized on 1.5% agarose–TAE (Tris-acetate-EDTA) gels stained with SYBR® Safe DNA gel stain (Invitrogen Merelbeke, Belgium), followed by purification using the High Pure PCR product purification kit (Roche, Vilvoorde, Belgium). Approximately 10 ng purified DNA was then used in a sequencing reaction with the BigDye terminator v3.1 sequencing kit (Applied Biosystems – Life Technologies, Gent, Belgium). Overlapping DNA fragments were sequenced in both directions using the corresponding forward and reverse primers used in the PCR amplification reaction. Full-length F-gene contigs were assembled from the partial sequences using SeqMan v 8.1.3 (DNASTAR, Madison, Wisconsin, USA). Genbank accession codes are summarized in .
The complete F-gene coding sequences were aligned with selected reference complete F-gene sequences (Genbank accession codes AJ880277, AY288992, AY288997, AY562989, AY562991, AY626267, AY626268, AY727883, AY741404, DQ097393, EF026583, EF564833, FJ410145, FJ410147, FJ772446, FJ772449, FJ772452, FJ772455, FJ772458, FJ772463, FJ772466, FJ772469, FJ772472, FJ772475, FJ772478, FJ772481, FJ772484, FJ772486, FJ772494, FJ772494, GQ429292, GU978777, and Z12111) (ClustalW using BioEdit v 7.1.3, http:///www.mbio.ncsu.edu/bioedit.html). These reference sequences included the most similar (BLASTn) complete F-gene coding sequences, and selected isolates representing the studied geographical area and the full genetic diversity of APMV-1. A neighbour-joining phylogenetic tree was calculated using Mega v4.0.2 (Tamura et al., Citation2007), using 1000 bootstrap pseudo-replications, pairwise deletion of gaps and missing data, and the Kimura two-parameter model. The previously described lineage-based nomenclature (Aldous et al., Citation2003; Cattoli et al., Citation2010) is used for identification of clades throughout this paper.
Results
Three viruses did not propagate in eggs, although they previously tested positive in virus isolation. All 12 isolates that were positive in virus isolation were confirmed in real-time RT-PCR, with cycle treshold values for the allantoic fluids ranging from 17.0 to 26.7. The complete F-gene coding sequence was only characterized for the 12 isolates that were APMV-1-positive, and the sequences were submitted to Genbank (accession codes presented in ). Three different cleavage site motifs, all velogenic, could be distinguished ().
A high degree of genetic diversity could be observed in the characterized viruses, which included members of lineages 7a, 7d, and 4b. At least five clusters could be identified from these 12 samples ().
Figure 1. Phylogenetic analysis of the complete F-gene coding sequence of Nigerian ND virus isolates. Bold underlined, characterized in the present study. Province of origin is indicated in brackets.
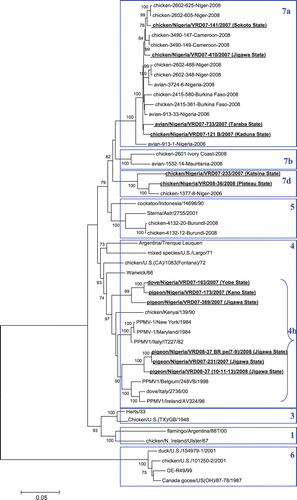
The presence of the recently identified lineage 7 could be confirmed in six samples from six different states (from Sokoto in the northwest to Taraba in eastern Nigeria), indicating a widespread circulation in local poultry. The associated epidemiological data () indicate variable levels of morbidity and mortality. Sub-lineage 7a was the most prevalent (). This sub-lineage contained at least two genetically distinct virus clusters (): one cluster with two chicken isolates from Sokoto and Jigawa states; and one cluster of poultry isolates in the Kaduna (chicken) and Taraba (unspecified poultry carcass from live bird market) states. We also detected two lineage 7d isolates from chickens in the Katsina and Plateau states ().
The highest number of samples was selected from Jigawa state (two samples in 2008, three samples in 2007). In spite of the limited sample size, our data show co-circulation in the same state of both lineages 7a and 4b of pigeon paramyxovirus type 1 (PPMV-1).
Lineage 4b viruses were detected in six samples from three states (). One cluster contains three closely related pigeon isolates (Jigawa state), while a related cluster contains one virus from dove (Yobe state), one virus from pigeon (Kano State) and one virus from pigeon (Jigawa state) (). Variable levels of clinical disease and mortality were associated with outbreaks of lineage 4b PPMV-1 viruses ().
Despite the close similarity of the viruses from each cluster within either lineage 4b or lineage 7, in some of these clusters the amino acid sequence of the cleavage site was variable. One cluster in lineage 7a contained chicken isolates VRD07-733 and VRD07-121B with respective cleavage site sequences RRQKRF and RRRKRF ( and ). Another cluster in lineage 4b contained two cleavage site motifs with equal numbers of basic amino acids RRQKRF and KRQKRF ( and ).
Discussion
ND is one of the most important livestock diseases affecting the subsistence of rural communities and the commercial poultry sector in Western Africa. Nevertheless, only limited information is available on the characterization of the ND virus strains circulating in this region. Two recent studies documented a large variety of genetic lineages circulating in that region (Snoeck et al., Citation2009; Cattoli et al., Citation2010). The survey of Snoeck and colleagues included samples from south-western Nigeria (period 2002 to 2007), showing circulation of lineages 1 to 3 nearly identical to vaccine strains in south-western Nigeria (Oyo, Ogun and Ekiti states) as well as one lineage 4 strain detected in a pigeon (Snoeck et al., Citation2009). In addition, and especially in rural poultry in northern Nigeria (Sokoto state), they documented divergent lineage 5 isolates (tentatively grouped in sub-lineages 5g, 5f and 5h) that were later included in a new lineage 7, specific for western and central Africa (Cattoli et al., Citation2010). A very recent paper, focused on live bird markets (Solomon et al., Citation2012), confirmed the circulation of these lineage 7 viruses (sub-lineages 5g and 5f in the terminology according to Snoeck et al. [Citation2009]) throughout Nigeria and showed a high degree of genetic diversification based on the phylogenetic analysis of a partial (314 base pair) F-gene sequence.
Our study focused on the rural poultry and meat pigeon sector in the central and north-eastern regions of Nigeria. Our findings confirm the previously documented presence of APMV-1 lineages 4b and 7 over a large geographic area in Nigeria. Our study targeted unvaccinated rural poultry. Not surprisingly, we did not detect the circulation of vaccine-related strains from lineages 1 to 3. Indeed, this survey was mainly based on clinical signs and we did not include commercial farms (the vaccinated part of the Nigerian poultry sector). This confirms the observations by Cattoli et al. (Citation2010) that did not find vaccine-related ND virus isolates from rural poultry. On the contrary, one-half of the isolates described by Snoeck et al. (Citation2009) in commercial farms and slaughterhouses were vaccine related. The latter study also documented two lineage 4b isolates in Nigeria (one from a pigeon and one from a parrot). The recorded mortality rates in our study (ranging from 19 to 36%) and the variety in the observed clinical signs together with the isolation of APMV-1 from birds without clinical signs () in one village are a strong indication of an endemic circulation of velogenic strains in the rural sector, jeopardizing the improvement of village chicken production and representing a constant threat for the industrial farms.
We observed two distinct clusters in sub-lineage 7a, and one additional cluster in sub-lineage 7d. This is consistent with the genetic diversification documented in live bird markets by Solomon et al. (Citation2012), and confirms the circulation of these viruses in backyard and small-scale chicken farms throughout Nigeria. All of these clusters contained isolates from multiple, non-neighbouring states. This, together with the high genetic diversity of lineage 7 APMV-1 isolates, is indicative of extensive and continued endemic circulation and genetic diversification as previously suggested (Cattoli et al., Citation2010; Solomon et al., Citation2012) All of these isolates have velogenic fusion-gene cleavage-site motifs, indicating their pathogenicity for poultry. This is confirmed by the epidemiological data reported here, with variable levels of morbidity and mortality documented during lineage 7 isolate outbreaks in rural poultry. Some genetically very similar isolates showed different cleavage site amino acid sequences. There is no indication from the epidemiological data that this resulted in differences in virulence.
In addition, lineage 4b viruses are circulating over a large geographic range with a considerable genetic diversity, and seem to persist exclusively in pigeons and feral doves while causing outbreaks of disease and mortality in these species. Lineage 4b viruses are referred to as PPMV-1 and can be distinguished antigenically from other APMV-1 (Meulemans et al., Citation2002; Aldous et al., Citation2004). Until recently they were mostly documented in the Middle East, Asia and Europe (Aldous et al., Citation2004). They have only been reported sporadically on the African continent (Ballouh et al., Citation1985; Snoeck et al., Citation2009). In the present study, three closely related isolates from the Jigawa state form a distinct cluster together with recent European PPMV-1. This cluster is associated with an outbreak of high mortality in domestic pigeons. A distinct cluster, more distant from contemporary European PPMV-1 isolates, regroups isolates from feral doves and domestic pigeons in the Yobe, Kano, and Jigawa states. This suggests at least two independent introductions of lineage 4b PPMV-1 in Nigeria. Apart from the direct impact on local producers relying on pigeon farming for meat production, sporadic introduction of PPMV-1 may result in velogenic disease in poultry (for example, Pearson et al., Citation1987; Kommers et al., Citation2001; Abolnik et al., Citation2008). Our data are the first to support endemic circulation of lineage 4b PPMV-1 in Western Africa. As suggested by data from South Africa (Abolnik et al., Citation2008), PPMV-1 may be circulating in other regions of the African continent.
All 12 virus isolation-positive samples in this study were real-time RT-PCR-positive. We did not observe the diagnostic problems with the M-gene real-time RT-PCR due to accumulation of mismatches in the probe binding site that was documented by Cattoli et al. (Citation2010) for some lineage 7 isolates. In that study, an accumulation of mismatches in the probe binding site was shown to result in false negative results in 7/16 virus isolation-positive samples from Niger and Mauretania. We did not observe this for the Nigerian isolates in this study (detected by M-gene real-time RT-PCR). However, high virus titres in the tested allantoic fluids may have masked inefficiency problems in the real-time RT-PCR in our study. Three samples that were previously virus isolation-positive did not contain any viable virus upon arrival at the VAR, possibly due to poor conservation. Unfortunately, insufficient starting material was left after our attempts to multiply the virus on eggs to confirm the presence of APMV-1 viral RNA in these isolates. These samples were thus excluded from the study.
In conclusion, the present study confirms the presence of the West African lineage 7 APMV-1 in Nigeria and documents its continuing circulation and diversification in rural poultry. In addition, we show evidence of endemic circulation of lineage 4b PPMV-1. Continuing diversification and circulation may impair validated molecular diagnostic tests and may have an influence on the efficacy of vaccination control (Miller et al., Citation2007). This indicates the importance of data generation and sequence sharing to monitor the efficacy of validated diagnostic tests and to monitor the emergence and diversification of new genetic lineages and sub-lineages.
Acknowledgements
Part of this study was conducted in the framework of the European Union network of excellence EPIZONE (FOODCT-2006-016236). The authors would like to express their gratitude to the OIE/FAO reference laboratory for avian influenza and ND at the IZSVe (Padova, Italy), the FAO in Rome (Gwenaelle Dauphin), and in particular the FAO office in Abuja (Nigeria) for logistic support. Orkun Ozhelvaci and Mireille Decaesstecker provided valued technical expertise. E. Obishakin was supported by the Master's programme “Interuniversity Programme in Molecular Biology” of the Flemish Interuniversity Council (Vlaamse Interuniversitaire Raad/VLIR).
Additional information
Notes on contributors
Steven Van Borm†
†These authors contributed equallyReferences
- Abolnik , C. , Gerdes , G.H. , Kitching , J. , Swanepoel , S. , Romito , M. and Bisschop , S.P. 2008 . Characterization of pigeon paramyxoviruses (Newcastle disease virus) isolated in South Africa from 2001 to 2006 . Onderstepoort Journal of Veterinary Research , 75 : 147 – 152 .
- Adene , D.F. and Oguntade , A.E. 2006 . The structure and importance of the commercial and village based poultry industry in Nigeria , Rome : FAO .
- Aldous , E.W. , Fuller , C.M. , Mynn , J.K. and Alexander , D.J. 2004 . A molecular epidemiological investigation of isolates of the variant avian paramyxovirus type 1 virus (PPMV-1) responsible for the 1978 to present panzootic in pigeons . Avian Pathology , 33 : 258 – 269 .
- Aldous , E.W. , Mynn , J.K. , Banks , J. and Alexander , D.J. 2003 . A molecular epidemiological study of avian paramyxovirus type 1 (Newcastle disease virus) isolates by phylogenetic analysis of a partial nucleotide sequence of the fusion protein gene . Avian Pathology , 32 : 239 – 256 .
- Alexander , D.J. 2000 . Newcastle disease and other avian paramyxoviruses . Scientific and Technical Review OIE , 19 : 443 – 462 .
- Alexander , D.J. 2003 . “ Newcastle disease ” . In Diseases of Poultry , Edited by: Saif , Y.M. , Barnes , H.J. , Glisson , J.R. , Fadly , A.M. , McDougald , L.R. and Swayne , D.E. 64 – 87 . Ames , IA : Iowa State Press .
- Ballouh , A. , Abu Elzein , E.M. and Elmubarak , A.K. 1985 . Outbreak of the pigeon paramyxovirus serotype 1 in the Sudan . The Veterinary Record , 116 : 375
- Bebay , C.E. 2006 . Première évaluation de la structure et de l'importance du secteur avicole commercial et familial en Afrique de l'Ouest. Synthèse des rapports nationaux , Rome : FAO/ECTAD/AGAP .
- Cattoli , G. , Fusaro , A. , Monne , I. , Molia , S. , Le Menach , A. , Maregeya , B. , Nchare , A. , Bangana , I. , Garba Maina , A. , N'Goran , K.J.-N. , Thiam , H. , Bezeid , O.E.M.A. , Salviato , A. , Nisi , R. , Terregino , C. and Capua , I. 2010 . Emergence of a new genetic lineage of Newcastle disease virus in West and Central Africa–implications for diagnosis and control . Veterinary Microbiology , 142 : 168 – 176 .
- Czegledi , A. , Ujvari , D. , Somogyi , E. , Wehmann , E. , Werner , O. and Lomniczi , B. 2006 . Third genome size category of avian paramyxovirus serotype 1 (Newcastle disease virus) and evolutionary implications . Virus Research , 120 : 36 – 48 .
- Kim , L.M. , Suarez , D.L. and Afonso , C.L. 2008 . Detection of a broad range of class I and II Newcastle disease viruses using a multiplex real-time reverse transcription polymerase chain reaction assay . Journal of Veterinary Diagnostic Investigation , 20 : 414 – 425 .
- Kommers , G.D. , King , D.J. , Seal , B.S. and Brown , C.C. 2001 . Virulence of pigeon-origin Newcastle disease virus isolates for domestic chickens . Avian Diseases , 45 : 906 – 921 .
- Lamb , R.A. , Collins , P.L. , Kolakofsky , D. , Melero , J.A. , Nagai , Y. , Oldstone , M.B.A. , Pringle , C.R. and Rima , B.K. 2005 . “ Family Paramyxoviridae ” . In Virus Taxonomy. Eighth report of the International Committee on Taxonomy of Viruses , Edited by: Fauquet , C.M. , Mayo , M.A. , Maniloff , J. , Desselberger , U. and Ball , L.A. 655 – 668 . Amsterdam : Elsevier .
- Lancaster , J.E. and Alexander , D.J. 1975 . Newcastle disease virus and spread. A review of some of the literature , Ottawa , , Canada : Canada Dept. of Agriculture (Ottawa) .
- Meulemans , G. , van den Berg , T.P. , Decaesstecker , M. and Boschmans , M. 2002 . Evolution of pigeon Newcastle disease virus strains . Avian Pathology , 31 : 515 – 519 .
- Miller , P.J. , King , D.J. , Afonso , C.L. and Suarez , D.L. 2007 . Antigenic differences among Newcastle disease virus strains of different genotypes used in vaccine formulation affect viral shedding after a virulent challenge . Vaccine , 25 : 7238 – 7246 .
- OIE 2008 . Manual of Diagnostic Tests and Vaccines for Terrestrial Animals , 6th edition , OIE World Organisation for Animal Health .
- Pearson , J.E. , Senne , D.A. , Alexander , D.J. , Taylor , W.D. , Peterson , L.A. and Russell , P.H. 1987 . Characterization of Newcastle disease virus (avian paramyxovirus-1) isolated from pigeons . Avian Diseases , 31 : 105 – 111 .
- Snoeck , C.J. , Ducatez , M.F. , Owoade , A.A. , Faleke , O.O. , Alkali , B.R. , Tahita , M.C. , Tarnagda , Z. , Ouedraogo , J.-B. , Maikano , I. , Mbah , P.O. , Kremer , J.R. and Muller , C.P. 2009 . Newcastle disease virus in West Africa: new virulent strains identified in non-commercial farms . Archives of Virology , 154 : 47 – 54 .
- Solomon , P. , Abolnik , C. , Joannis , T.M. , & Bisschop , S. 2012 . Virulent Newcastle disease virus in Nigeria: identification of a new clade of sub-lineage 5f from livebird markets . Virus Genes , 44 98 – 103 .
- Tamura , K. , Dudley , J. , Nei , M. and Kumar , S. 2007 . MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0 . Molecular Biology and Evolution , 24 : 1596 – 1599 .