Abstract
Chicken anaemia virus (CAV) is an economically important pathogen of chickens with worldwide distribution. CAV is the causative agent of chicken anaemia disease, causing severe anaemia, lymphoid atrophy and immunosuppression in young birds. In the present study, the genetic variation of circulating CAVs in west Azerbaijan broiler farms was investigated and compared with CAVs from other countries. Extracted viral DNA from livers of chickens positive for CAV (46 out of 100) was used and a fragment of the VP1 gene 1390 base pairs in size was amplified. The purified products were subjected to restriction fragment-length polymorphism (RFLP) using HinfI endonuclease and nucleotide sequencing. Four different RFLP patterns were identified from all examined CAV DNAs. Sequence analysis of the VP1 gene of isolated CAV viruses revealed a high genetic distance (0.5 to 4.7%) among CAV isolates. Phylogenetic analysis showed that CAVs isolated from Iranian poultry farms clustered with CAVs isolated from different parts of the world. It was concluded that the circulating CAVs in broiler farms of west Azerbaijan had a high genomic variation.
Introduction
Chicken anaemia virus (CAV), a member of the genus Gyrovirus (Pringle, Citation1999) of the Circoviridae family with worldwide distribution, was first recognized in Japan by Yuasa (Schat, Citation2003). The virus is a small, non-enveloped, icosahedron with an average diameter of 25 to 26.5 nm consisting of a negative-sense, single-stranded circular DNA genome (Schat, Citation2003). The CAV genome size is of 2.3 kb consisting of three partially overlapping open reading frames encoding three main proteins of the virus. VP1 is the major viral structural protein of CAV with the highest variability of the three overlapping open reading frames; VP2 is a scaffolding protein with dual-specificity protein phosphatase (Peters et al., Citation2002); and VP3 is a non-structural protein named apoptin (13.6 kDa) that is able to induce apoptosis (Schat, Citation2003). VP1 and VP2 are the main targets of neutralizing antibodies (Noteborn et al., Citation1992).
CAV causes the acute and immunosuppressive disease chicken infectious anaemia (MacLachlan & Dubovi, Citation2011) in unprotected young chickens 2 to 4 weeks old, characterized by aplastic anaemia and generalized lymphoid atrophy (Schat, Citation2003). Signs such as haemorrhages in leg and chest muscles, focal necrosis in the liver, ulcerative erosions in the gizzard, and necrosis of wing skin can be observed in infected birds with CAV (Smyth et al., Citation1993). In the field, CAV infection seems to cause few signs of disease; however, dual infections are more serious (McNulty, Citation1991; Dhama et al., Citation2002). The mortality due to CAV in a flock is generally low but can be high due to secondary complications; the virus therefore seems to play a key role in the aetiology of several multifactorial diseases (Pope, Citation1991; Todd, Citation2000).
Conventional diagnostic methods such as histopathological examination, electron microscopy and routine virus isolation techniques are relatively expensive, time consuming and unable to detect CAV strains that fail to replicate in MSB1 or other cell lines (Saini & Dandapat, Citation2009). Different molecular techniques such as polymerase chain reaction (PCR), hybridization assay, restriction enzyme analysis and sequencing have therefore been developed for CAV detection and characterization (Noteborn et al., Citation1992; Saini & Dandapat, Citation2009). VP1, the major and the sole structural protein of CAV with the highest variability and also liable for intergenotype and intersubtype recombination events (He et al., Citation2007; Eltahir et al., Citation2011a), is the most appropriate component of the virus for molecular characterization.
The first occurrence of CAV infection among Iranian broiler flocks was reported by Toroghi et al. in 2003. Farhoodi et al. (Citation2007) reported widespread distribution of CAV in three provinces including Tehran, Isfahan and Khorasan. To the best of our knowledge there are no CAV nucleotide sequence data from Iranian CAV isolates. In the present study, CAV isolates from broiler chickens of west Azerbaijan were characterized and compared with CAV strains circulating in the other parts of the world by PCR, restriction fragment length polymorphism (RFLP) and nucleotide sequencing. Phylogenetic analysis showed that the isolated viruses were clustered differently from each other, one of them grouped separately from all other CAVs reported from other countries; further sequence analysis showed that this isolate was a recombinant.
Materials and Methods
Sample collection and virus detection
A number of 100 chicken liver tissues were collected randomly from broiler chicken slaughterhouses across west Azerbaijan province, Iran. The health status of examined chickens was not recorded. The livers were removed aseptically, transferred in ice to the laboratory and were frozen at −20°C for subsequent DNA extraction.
DNA extraction
Liver tissues were crushed in sterile glass Petri dishes with phosphate-buffered saline buffer. The crushed tissue was centrifuged at 736×g for 10 sec and 100 µl supernatant was used for DNA extraction. The DNA extraction was performed using a High Pure Viral genomic extraction kit (Roche, Mannheim, Germany) according to the kit manufacturer's instruction.
Amplification of the VP1 gene
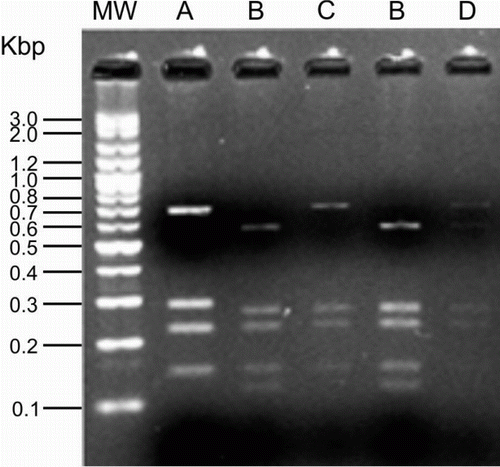
For amplification of the VP1 gene of CAV, two primers targeting 1390 base pairs (bp) comprising the whole VP1 gene of CAV were used. The forward primer (5′-AGC CGA CCC CGA ACC GCA AGA A-3′) was described by Farhoodi et al. (Citation2007) and the reverse primer (5′-TCA GGG CTG CGT CCC CCA GTA CA-3′) was previously described by Mohamed (Citation2010). The PCR was carried out in 25 µl mixtures containing 50 mM each of dATP, dTTP, dGTP and dCTP, 0.5 mM each primers 2.5 µl of 10× PCR buffer (Cinnagen, Tehran, Iran), 2 mM magnesium chloride, 2.5 U Taq DNA polymerase (Cinnagen) and 50 to 100 ng extracted DNA as template. The cycling conditions for amplification were performed using an initial incubation at 94°C for 2 min and 35 cycles of incubation at 94°C for 45 sec, 62°C for 45 sec and 72°C for 110 sec followed by a final extension at 72°C for 5 min. The resultant PCR products were separated in a 1.5% agarose gel and the gel photographed using ultraviolet trans-illumination.
PCR products purification from agarose gel
PCR products were purified from agarose gel using a PCR purification kit (GF1-Ambiclean kit; Vivantis, Selangor, Malaysia) according to the manufacturer's instructions.
Restriction fragment length polymorphism
Purified PCR products were digested with the HinfI restriction endonuclease, which was chosen based on a simulated restriction map of amplified PCR products using BioEdit software (Hall, Citation1999). Digestion reaction was carried out in 15 µl mixtures containing 5 μl purified PCR product, 5U HinfI endonuclease (Cinnagen) and 1.5 µl HinfI buffer. The obtained mixtures were incubated at 37°C for 2 h. After digestion, the resultant DNA fragments were separated in 1.5% agarose gel and photographed using ultraviolet trans-illumination.
DNA sequencing
The VP1 gene purified PCR products from four CAV isolates with different RFLP patterns along with both forward and reverse primers were sent to CinnaClone (Karaj, Iran) for nucleotide sequencing.
Nucleotide sequence analysis
DNA sequences of the VP1 gene of four CAV isolates were aligned using ClustalW (Thompson et al., Citation1994) and compared with 20 foreign CAV isolates retrieved from the GenBank database. The sequence names of the retrieved CAV isolates, their GenBank accession numbers and the country of origin are presented in . Two retrieved sequences were from strains Cuxhaven-1 and 26P4 that are used for vaccination on Iranian poultry farms, especially breeder farms. The phylogenetic tree was generated using the maximum-likelihood method based on the Kimura two-parameter model. Evolutionary divergence between sequences was analysed using the Kimura two-parameter model (Kimura, Citation1980), which corrects for multiple hits, taking into account transitional and transversional substitution rates, while assuming that the four nucleotide frequencies are the same and that rates of substitution do not vary among sites. Genetic distances were estimated by computing the proportion of nucleotide differences between each pair of nucleotide sequences. Phylogenetic and evolutionary divergence analysis involved 21 nucleotide sequences. All positions containing gaps and missing data were eliminated. There were 1285 positions in total in the final dataset. Evolutionary analyses were conducted in MEGA5 (Tamura et al., Citation2007).
Table 1. CAV nucleotide sequence names, Genbank accession numbers and the country of origin for the sequences used in the present study.
Similarity plot and breakpoint analysis for identification of putative recombinant sequence
The aligned sequences of the VP1 gene of CAV isolates in the present study with reference VP1 sequences from Genbank were introduced into SimPlot version 3.5.1 to identify probable recombination sites (Lole et al., Citation1999). The VP1 gene of Cuxhaven strain (the strain currently in use in Iranian poultry farms) was used as a query to determine the potential recombinant sequence. Bootscanning analysis was performed in Simplot to identify and map the putative recombinant VP1 sequences (Salminen et al., Citation1995).
Results
Amplification of 1.4 kb of the VP1 gene and purification of PCR products
All chicken livers collected from broiler slaughterhouses were subjected to DNA extraction. A 1.4 kb fragment of nearly complete VP1 gene was amplified from 46 DNAs isolated from liver samples by PCR. All amplified PCR products were cleaned up from the gel and were subjected to RFLP and nucleotide sequencing.
Restriction endonuclease digestion of PCR products
The RFLP patterns generated from VP1 gene PCR products of CAV isolates by digestion with HinfI are shown in . Examining all positive liver samples for CAV, a total number of four different RFLP patterns (A, B, C and D) were generated. CAVs with RFLP pattern B had the highest frequency (55%) and all other patterns had almost similar frequencies.
Nucleotide sequence analysis of the VP1 gene of CAV isolate with different RFLP patterns
Comparison of the nucleotide sequences of the VP1genes of four CAV isolates with different RFLP patterns with the VP1 nucleotide sequences of CAVs from other countries revealed distinct difference not only among VP1 genes of Iranian CAVs but also between Iranian and CAVs from other countries. The VP1 gene of Ur-41 had the highest genetic distance for all VP1 genes of CAVs examined in this study and CAVs from other countries (3.5 to 4.7%). Among four CAVs from Iran, Ur-19 was most similar to CAVs isolated from other countries with 0.5 to 2.2% genetic distance. Ur-24 and Ur-32 were more similar (99.99%) to each other than to Ur-19 and Ur-41 (). The phylogenetic tree constructed showed similar results as nucleotide divergence analysis. Based on the phylogenetic tree, Ur-41 clustered separately from all CAVs examined in the present study, indicating that this isolate was highly different from other CAVs. Ur-24 and UR-32 clustered very close to each other, mostly with isolates from the USA. Ur-19, with the lowest genetic distance compared with the other CAVs, clustered mostly with CAVs isolated from Asian and European countries ().
Figure 2. Molecular phylogenetic analysis of 21 nucleotide sequences of CAV VP1 gene by the maximum-likelihood method.
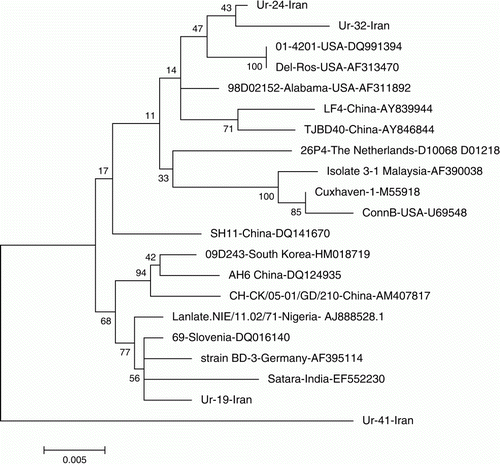
Table 2. Estimates of evolutionary divergence between nucleotide sequences of the CAV VP1 gene.
Identification of putative recombinant sequence
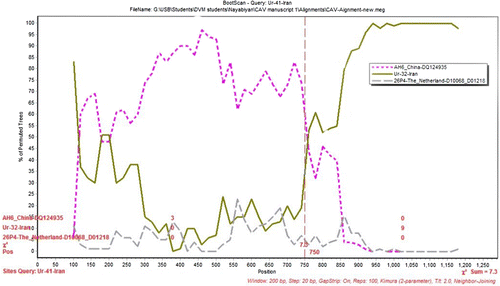
To identify potential mosaic among the 21 CAV sequences, similarity plots and bootscan analyses were performed. The results showed that Ur-41-Iran isolate was a natural recombinant that was descended from two putative parents (AH6 strain from China and Ur-32-Iran isolate from Iran) in the VP1 gene. The potential breakpoints analyses based on the VP1 gene of Ur-41-Iran revealed that the single breakpoint was located in a parsimonious region from 720 to 760 with maximization of X2 using the program Simplot ().
Figure 3. The results of bootscanning of Ur-41-Iran. The y axis gives the percentage of permutated trees using a sliding window of 200 bp wide centred on the position plotted, with a step size between plots of 20 bp. AH6, Ur-32-Iran and 26P4 strains were used as the outgroup to determine the breakpoint.
Discussion
CAV is an economically important avian pathogen worldwide due to its highly immunosuppressive nature. In Iran, there is not much information on CAVs circulating in the poultry population of the country. Therefore, in the present study, molecular characterization of CAV isolates including RFLP analysis and nucleotide sequencing was undertaken to investigate the variation among examined CAV isolates, aiming to establish a suitable disease control approach in the poultry industry.
In the present report PCR-RFLP and nucleotide sequencing procedures were used to detect and differentiate CAV DNA in tissue extracts from infected birds. The variation among the examined Iranian CAV isolates ranged from 0.5 to 4.7%, which was similar to variation among Indian CAV isolates (0.5 to 4.0%) reported by Natesan et al. (Citation2006). In another report by Oluwayelu et al. (Citation2008), nucleotide diversity obtained for the Nigerian commercial and backyard chicken strains was 6% and 4%, respectively. Based on phylogenetic analysis, Chinese CAV isolates were classified into four distinct sequence groups (A to D), indicating that four genetically different CAV clusters are circulating in China (Eltahir et al., 2011). The phylogenetic analysis of Iranian CAVs showed genetic variability among field isolates of CAV and their nucleotide distances from CAV vaccine strains (Cuxhaven-1 and 26P4). The results also revealed that the Iranian CAV isolates showed limited variations with isolates from the other countries except for the recombinant isolate. CAV intergenotype recombination break points in the VP1 region have been reported previously (He et al., Citation2007). In addition to intergenotype recombination, intersubtype recombination events in CAV genome sequences were also reported in CAV subgroup A1 and A2 sequences and these recombination events were found in the VP1 gene (Eltahir et al., 2011). It can be concluded that genomic recombination events in the CAV genome most probably occur in the VP1 variable region.
However, no significant antigenic or pathogenic difference was reported among the CAV isolates in the past and CAV was known as a much-conserved virus of one serotype (McNulty et al., Citation1990) with several genetic groups (Islam et al., Citation2002). Genotyping of CAV based on RFLP and even sequencing will therefore be of limited benefit for establishing suitable prevention and control strategies for CAV infection in the poultry industry.
The widespread CAV in Iranian broiler poultry farms as reported (Toroghi et al., Citation2003; Farhoodi et al., Citation2007) and in the current study highlights the importance of this avian pathogen as a suppressor of the immune system of infected chickens and increases the risk of secondary infections such as infectious bronchitis, Newcastle disease, colibacilosis and other diseases. Epidemiology of CAV infection should therefore be investigated in all provinces to advise suitable prevention strategies for this economically important avian pathogen and to strengthen the necessity for providing adequate immunity of breeder flocks as a means of avoiding the infection in their progeny.
Acknowledgements
This research was supported by the Dean of Research, Urmia University. The authors thank Dr F. Zobairi for her help with methods.
References
- Dhama , K. , Kataria , J.M. , Dash , B.B. , Senthilkumar , N. and Tomar , S. 2002 . Chicken infectious anaemia (CIA): a review . Indian Journal of Comparative Microbiology, Immunology and Infectious Diseases , 23 : 1 – 15 .
- Eltahir , Y.M. , Qian , K. , Jin , W. and Qin , A. 2011a . Analysis of chicken anemia virus genome: evidence of intersubtype recombination . Virology Journal , 8 : 512 doi: 10.1186/1743-422X-8-512
- Eltahir , Y.M. , Qian , K. , Jin , W. , Wang , P. and Qin , A. 2011b . Molecular epidemiology of chicken anemia virus in commercial farms in China . Virology Journal , 8 : 145 doi: 10.1186/1743-422X-8-145
- Farhoodi , M. , Toroghi , R. , Bassami , M.R. and Kianizadeh , M. 2007 . Chicken infectious anaemia virus infection among broiler chicken flocks in Iran . Archives of Razi Institute , 62 : 1 – 6 .
- Hall , T.A. 1999 . Bioedit: a user-friendly biological sequence alignment editor and analysis program for windows 95/98/nt . Nucleic Acids Symposium Series , 41 : 95 – 98 .
- He , C.Q. , Ding , N.Z. , Fan , W. , Wu , Y.H. , Li , J.P. and Li , Y.L. 2007 . Identification of chicken anemia virus putative intergenotype recombinants . Virology , 366 : 1 – 7 . doi: 10.1016/j.virol.2007.06.007
- Islam , M.R. , Johne , R. , Raue , R. , Todd , D. and Muller , H. 2002 . Sequence analysis of the full-length cloned DNA of a chicken anaemia virus (CAV) strain from Bangladesh: evidence for genetic grouping of CAV strains based on the deduced VP1 amino acid sequences . Journal of Veterinary Medicine Series B, Infectious Diseases and Veterinary Public Health , 49 : 332 – 337 . doi: 10.1046/j.1439-0450.2002.00581.x
- Kimura , M. 1980 . A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences . Journal of Molecular Evolution , 16 : 111 – 120 . doi: 10.1007/BF01731581
- Lole , K.S. , Bollinger , R.C. , Paranjape , R.S. , Gadkari , D. , Kulkarni , S.S. , Novak , N.G. , Ingersoll , R. , Sheppard , H.W. and Ray , S.C. 1999 . Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination . Journal of Virology , 73 : 152 – 160 .
- MacLachlan , N.J. & Dubovi , E.J. 2011 . Fenner's Veterinary Virology. , 4th edn 237 – 242 . Amsterdam , Elsevier .
- McNulty , M.S. 1991 . Chicken anaemia agent: a review . Avian Pathology , 20 : 187 – 203 . doi: 10.1080/03079459108418756
- McNulty , M.S. , Connor , T.J. , McNeilly , F. , McLoughlin , M.F. and Kirkpatrick , K.S. 1990 . Preliminary characterisation of isolates of chicken anaemia agent from the United Kingdom . Avian Pathology , 19 : 67 – 73 . doi: 10.1080/03079459008418657
- Mohamed , M.A. 2010 . Chicken infectious anemia status in commercial broiler chickens flocks in Assiut-Upper Egypt: occurrence, molecular analysis using PCR-RFLP and apoptosis effect on affected tissues . International Journal of Poultry Science , 9 : 591 – 598 . doi: 10.3923/ijps.2010.591.598
- Natesan , S. , Kataria , J.M. , Dhama , K. , Rahul , S. and Baradhwaj , N. 2006 . Biological and molecular characterisation of chicken anaemia virus isolates of Indian origin . Virus Research , 118 : 78 – 86 . doi: 10.1016/j.virusres.2005.11.017
- Noteborn , M.H. , Kranenburg , O. , Zantema , A. , Koch , G. , de Boer , G.F. and van der Eb , A.J. 1992 . Transcription of the chicken anemia virus (CAV) genome and synthesis of its 52-kda protein . Gene , 118 : 267 – 271 . doi: 10.1016/0378-1119(92)90198-X
- Oluwayelu , D.O. , Todd , D. and Olaleye , O.D. 2008 . Sequence and phylogenetic analysis of chicken anaemia virus obtained from backyard and commercial chickens in Nigeria . Onderstepoort Journal of Veterinary Research , 75 : 353 – 357 . doi: 10.4102/ojvr.v75i4.111
- Peters , M.A. , Jackson , D.C. , Crabb , B.S. and Browning , G.F. 2002 . Chicken anemia virus vp2 is a novel dual specificity protein phosphatase . The Journal of Biological Chemistry , 277 : 39566 – 39573 . doi: 10.1074/jbc.M201752200
- Pope , C.R. 1991 . Chicken anemia agent . Veterinary Immunology and Immunopathology , 30 : 51 – 65 . doi: 10.1016/0165-2427(91)90008-Z
- Pringle , C.R. 1999 . Virus taxonomy at the xith international congress of virology, Sydney, Australia, 1999 . Archives of Virology , 144 : 2065 – 2070 . doi: 10.1007/s007050050728
- Saini , N.S. and Dandapat , A. 2009 . Diagnosis and molecular characterisation of chicken anaemia virus . Veterinary World , 2 : 156 – 160 .
- Salminen , M.O. , Carr , J.K. , Burke , D.S. and McCutchan , F.E. 1995 . Identification of breakpoints in intergenotypic recombinants of HIV type 1 by bootscanning . AIDS Research and Human Retroviruses , 11 : 1423 – 1425 . doi: 10.1089/aid.1995.11.1423
- Schat , K.A. 2003 . “ Chicken infectious anemia ” . In Diseases of Poultry , Edited by: Saif , Y.M. , Barnes , H.J. , Glisson , J.R. , Fadly , A.M. , McDougald , L.R. and Swayne , D.E. 182 – 202 . Ames , IA : Iowa State University Press .
- Smyth , J.A. , Moffett , D.A. , McNulty , M.S. , Todd , D. and Mackie , D.P. 1993 . A sequential histopathologic and immunocytochemical study of chicken anemia virus infection at one day of age . Avian Diseases , 37 : 324 – 338 . doi: 10.2307/1591656
- Tamura , K. , Dudley , J. , Nei , M. and Kumar , S. 2007 . Mega4: molecular evolutionary genetics analysis (mega) software version 4.0 . Molecular Biology and Evolution , 24 : 1596 – 1599 . doi: 10.1093/molbev/msm092
- Thompson , J.D. , Higgins , D.G. and Gibson , T.J. 1994 . Clustal W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice . Nucleic Acids Research , 22 : 4673 – 4680 . doi: 10.1093/nar/22.22.4673
- Todd , D. 2000 . Circoviruses: immunosuppressive threats to avian species: a review . Avian Pathology , 29 : 373 – 394 . doi: 10.1080/030794500750047126
- Toroghi , R. , Shoshtari , A.H. , Charkhkar , S. & Neyazi , M.H. 2003 . The first report of chicken infectious anemia occurrence among iranian broiler flocks . 13th Iranian Veterinary Congress 240 . Tehran Iranian Veterinary Association