Abstract
Mycoplasma synoviae infections result in significant economic losses in the chicken and turkey industries. A commercially available live temperature-sensitive (ts +) vaccine strain MS-H has been found to be effective in controlling M. synoviae infections in commercial layer and broiler breeder farms in various countries, including Australia. Detection and differentiation of MS-H from field strains (ts −) and from ts − MS-H reisolates in vaccinated flocks is vital in routine flock status monitoring. At present microtitration is the only available technique to determine the ts phenotype of M. synoviae. This technique is time consuming and not amenable to automation. In the present study, a quantitative real-time polymerase chain reaction (Q-PCR) was combined with simultaneous culturing of M. synoviae at two different temperatures (33°C and 39.5°C) to determine the ts phenotype of 22 Australian M. synoviae strains/isolates. The M. synoviae type strain WVU-1853 was also included for comparison. A ratio of the copy numbers of the variable lipoprotein haemagglutinin (vlhA) gene at the two temperatures was calculated and a cut-off value was determined and used to delineate the ts phenotype. In all M. synoviae strains/isolates tested in this study, the ts phenotype determined using Q-PCR was in agreement with that determined using conventional microtitration. Combination of Q-PCR with differential growth at two different temperatures is a rapid, reliable and accurate technique that could be used as an effective tool in laboratories actively involved in ts phenotyping of M. synoviae strains/isolates.
Introduction
Mycoplasma synoviae (MS) causes airsacculitis and infectious synovitis in poultry (Kleven & Ferguson-Noel, Citation2008) and inflicts considerable economic losses to the poultry industry due to carcass condemnation, culling of lame birds and inferior eggshell quality (Feberwee et al., Citation2009; Feberwee & Landman, Citation2010). Vaccination is an effective means to control the disease when biosecurity measures fail to prevent infection of poultry flocks. The temperature-sensitive (ts +) M. synoviae strain MS-H (Vaxsafe MS; Bioproperties Pty Ltd, Ringwood, Victoria, Australia) is the only commercially available live M. synoviae vaccine and is used in various countries, including Australia.
Where investigated, the MS-H vaccine was readily distinguishable from most, if not all, field strains of M. synoviae isolated from several countries by examination of the conserved region of the vlhA gene (Benčina et al., Citation2001; Hong et al., Citation2004; Jeffery et al., Citation2007; Hammond et al., Citation2009; Harada et al., Citation2009; Ogino et al., Citation2011; Ramírez et al., Citation2011). However, in Australia, most M. synoviae field strains are indistinguishable from MS-H using this method (Jeffery et al., Citation2007). Restriction fragment length polymorphism of genomic DNA has been used to distinguish between MS-H and M. synoviae field strains; however, this technique is time consuming and results may be difficult to interpret. In addition, restriction fragment length polymorphism does not differentiate MS-H from its ts − reisolates. Unlike MS-H, ts − MS-H clones were associated with minor changes in tracheal mucosa (Noormohammadi et al., Citation2003), and therefore it is important to differentiate the ts + MS-H from its ts − MSH clones. Currently, microtitration at two different temperatures (33°C and 39.5°C) is the only available technique to differentiate ts + MS-H from field strains or ts − MS-H reisolates (Morrow et al., Citation1998). But the conventional microtitration method is still time consuming, laborious and not amenable to automation. In this study, a quantitative real-time polymerase chain reaction (Q-PCR) in combination with growth at two different temperatures (33°C and 39.5°C) was used to determine the ts phenotype of M. synoviae strains/isolates.
Materials and Methods
M. synoviae strains/isolates and growth media
A total of 23 M. synoviae strains/isolates were used in this study. They included MS-H vaccine and its parent strain 86079/7NS, and a number of M. synoviae isolates that were identified as MS-H related by restriction fragment length polymorphism of genomic DNA (Markham et al., Citation1998b; Noormohammadi et al., Citation2003) (). All strains were grown in mycoplasma broth (MB) (Whithear, Citation1993) as described earlier (Jones et al., Citation2006).
Table 1. Origin of the M. synoviae strains/isolates used and their temperature-sensitivity (ts) phenotypes as determined by microtitration and vlhA Q-PCR.
Microtitration of M. synoviae strains for ts phenotyping
Microtitration of M. synoviae strains/isolates was performed as described earlier (Morrow et al., Citation1998). Briefly, 225 µl MB was added to each well of a 96-well microtitre plate (Nunc™, Roskilde, Denmark) and the wells of the first column were each inoculated with 25 µl of one of the M. synoviae strain/isolate cultures. The contents of the wells were mixed at least five times followed by transfer of 25 µl to the wells of the next column using a multichannel pipette. The transfer of diluted cultures to the wells of the next column was repeated for all columns except for column 12, which was used as the sterility (negative) control and contained MB only. The plate was sealed with an adhesive cover. A duplicate plate was prepared using the same procedure, with one plate incubated at 33°C and the other at 39.5°C for 2 weeks. Any well with a noticeable change in MB colour from red to yellow/orange was considered as positive growth and the colour-changing units per millilitre (CCU/ml) of each strain/isolate at each temperature were calculated using a most probable number table (Meynell & Meynell, Citation1970). Temperature-sensitive cultures were defined as those having a decrease in titre ≥103 CCU/ml when grown at 39.5°C compared with 33°C.
Preparation of pH gradient
A series of MB with different pH values (8.0 to 6.3) was prepared to aid in harvesting the mycoplasma cultures at approximately the same growth phase (Supplementary material, Figure S1, online only). One-millilitre aliquots of MB in Eppendorf tubes were mixed with 0, 2, 4, 6, 8, 10, 12, 14, 16, 18, 20 and 22 µl filter-sterilized 0.5 M hydrochloric acid solution to develop a pH gradient from 8.0 to 6.3 in MB tubes producing a range in colour from deep red to yellow caused by phenol red indicator in the broth media.
Growing M. synoviae strains for quantitative real-time PCR
One-millilitre volumes of MB in Eppendorf tubes were inoculated with 1/5, 1/10 and 1/20 dilution of M. synoviae cultures in duplicate, with one tube incubated at 33°C and the other at 39.5°C. Different dilutions of the same inocula were used to provide flexibility for the time of harvest, but a single dilution was selected for each culture and used for subsequent experiments. When either of the cultures (33°C or 39.5°C) showed colour change to orange–yellow (approximately pH 6.8 as determined by comparison with colour gradient), both tubes were harvested and either stored at −70°C for future extraction or immediately subjected to genomic DNA extraction as described below.
Ten-millilitre volumes of MS-H and 86079/7NS were also grown at 33°C and 39.5°C for up to 4 days to examine the effect of pH on vlhA Q-PCR results. One-millilitre aliquots of cultures were removed after each 24 h interval, their pH was determined and they were then subjected to DNA extraction.
Genomic DNA extraction
To extract genomic DNA, 500 µl of each culture was centrifuged at 14,000×g for 3 min. The cell pellet was resuspended in 500 µl RLT lysis buffer (Qiagen, Chadstone Centre, Victoria, Australia) containing 1% β-mercaptoethanol and incubated at 4°C overnight. Then 15 µl of Qiaex II and 300 µl of 70% ethanol were added to the cell suspension in lysis buffer, mixed and the suspension was loaded into a multispin MSK-11 column (Axygen, Union City, California, USA). The columns were centrifuged for 30 sec at 10,000×g at room temperature and the flow-through was discarded. The columns were washed once with 600 µl RW1 buffer (Qiagen) and twice with 500 µl RPE buffer (Qiagen) followed by centrifugation for 30 sec at 10,000×g and for 90 sec at 18,000×g. Finally, 40 µl diethyl pyrocarbonate-treated water was added to the columns, and after incubation for 5 min at room temperature the genomic DNA was eluted by centrifugation for 60 sec at 10,000×g. The eluted DNA was immediately used in Q-PCR or stored at −20°C for future use.
Preparation of standard curve for quantitative real-time PCR
M. synoviae vlhA gene (vlhA3; GenBank accession number AF085698) was PCR amplified using primers PCR-F (GATGCGTAAAATAAAAGGATTT) and PCR-R (ATGTTTTTGGTTTTATTATTATTA). Purified PCR product was cloned at the SmaI site of pUC18 plasmid (GenScript, Piscataway, New Jersey, USA) (Noormohammadi et al., Citation2000) and the final construct (5047 base pairs) was used to generate a standard curve for vlhA Q-PCR. The plasmid DNA concentration, measured using spectrophotometry (NanoDrop® ND-1000; Wilmington, Delaware, USA) at 260 nm, was adjusted to 3.5 ng/µl. Serial 10-fold dilutions of plasmid DNA were prepared in nuclease-free water for each experiment. The plasmid copy number in each preparation ranged from 109 to 101 copies as calculated using the following formula provided by URI Genomics & Sequencing Center (http://www.uri.edu/research/gsc/resources/cndna.html):
vlhA quantitative real-time PCR
For Q-PCR, oligonucleotide primers Link-F and MS-Cons-R (Jeffery et al., Citation2007) were used to amplify the single-copy conserved region at the 5′ end of the vlhA gene. A 25 µl reaction mixture consisted of 250 µM of each of dATP, dGTP, dCTP and dTTP (Promega, Alexandria, New South Wales, Australia), 2 mM MgSO4, 1 µl each of 25 µM forward and reverse oligonuncleotides, 8 µM SYTO 9 green fluorescent nucleic acid stain (Invitrogen, Mount Waverley, Victoria, Australia), 1 u of Platinum® Taq DNA polymerase high fidelity (Invitrogen), 1× high-fidelity PCR buffer and 3 µl extracted genomic DNA or plasmid DNA from each standard dilution. Amplification of DNA was performed in a Rotor-Gene 6000 thermal cycler (Corbett Life Science, Mortlake, New South Wales, Australia) using thermocycling conditions of one cycle of 96°C for 2 min, and 40 cycles of 96°C for 15 sec, 54°C for 15 sec and 68°C for 20 sec. Each DNA sample including each dilution of plasmid DNA used for generation of standard curve was analysed in triplicate in each experiment. Q-PCR results were analysed using the software Rotor-Gene 1.7.27 (Corbett Life Science, Mortlake, New South Wales, Australia), and algorithms provided. For each strain, vlhA copy numbers per reaction at 33°C and 39.5°C were obtained and averaged.
Results
Temperature-sensitive phenotypes determined using microtitration
The temperature-sensitive phenotype of 23 M. synoviae strains/isolates was determined using conventional microtitration at 33°C and 39.5°C. The MS-H vaccine and its parent strain 86079/7NS were included in each microtitration experiment to serve as ts + and ts − controls, respectively (). Thirteen M. synoviae strains/isolates were found to be ts − while 10 were ts +. The difference in most probable number obtained at the two different temperatures ranged from 3 to 8 logs in CCU/ml for the ts + strains, and from 0 to 2 logs for ts − strains/isolates. All ts + and ts − strains grew at 33°C whilst some ts + strains/isolates did not show any growth at 39.5°C (and therefore their titre was considered as 0) while some produced only a slight colour change up to the first or second column of the microtitration plates. Most ts − strains/isolates changed the medium colour to yellow at 39.5°C, while strain 86079/7NS produced an orange rather than yellow medium at 39.5°C.
Reproducibility of the vlhA quantitative real-time PCR
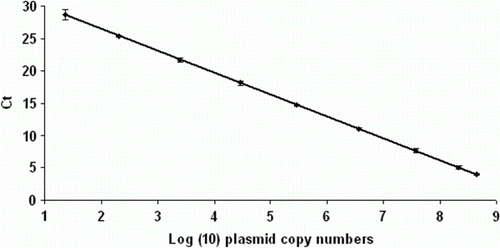
The standard curves generated using 10-fold serial dilutions of the pUC18 plasmid harbouring the vlhA3 gene had an average coefficient of determination (R 2) of 0.983 and an average efficiency of 85% (). The reproducibility of the assay was assessed by examination of variation in cycle threshold values. An average standard deviation of 2.23 was obtained in five independent experiments using pUC18 plasmid diluted from 109 to 101 copies per reaction.
Optimum pH to harvest M. synoviae cultures for determination of ts phenotype using vlhA quantitative real-time PCR
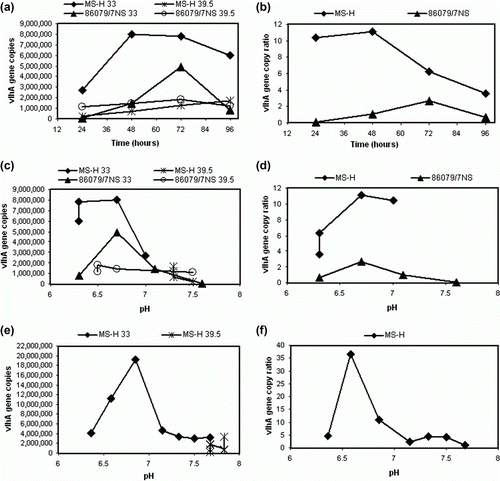
The vlhA gene copy numbers were determined using Q-PCR for MS-H and 86079/7NS cultures incubated at 33°C and 39.5°C and harvested at 24-h intervals to 96 h (a). At each time point, the pH was also determined and readings were plotted against vlhA copy numbers (c). At 33°C, MS-H and 86079/7NS grew to their highest titre (as determined by the vlhA gene copy number) at 48 and 72 h after inoculation, respectively. When plotted against pH, the highest vlhA gene copy number at 33°C was observed at an approximate pH of 6.8 for both MS-H and 86079/7NS, while at 39.5°C the vlhA gene copy number remained relatively steady at different pH values tested (a,c). A ratio was calculated by dividing the average vlhA copy number of the culture grown at 33°C by that at 39.5°C. The highest 33/39.5°C vlhA gene copy number ratios for MS-H and 86079/7NS cultures were found at 48 and 72 h after inoculation, respectively, with the ratio decreasing for both cultures with longer incubation (b). When plotted against pH, the highest 33/39.5°C ratio was found at an approximate pH of 6.8 (d). The 33/39.5°C vlhA gene copy number ratios for 86079/7NS always remained lower than those of MS-H whether plotted against time or pH (b and d respectively). Therefore, harvesting cultures at an approximate pH of 6.8 was found to be optimal for better discrimination of the ts phenotype. In a separate experiment, the 33°C and 39.5°C MS-H cultures were harvested at different pH irrespective of the time (e) and 33/39.5°C vlhA gene copy number ratios were determined (f). The results were consistent with those described above. The 33/39.5°C vlhA gene copy number ratio was highest at an approximate pH of 6.8 and then gradually decreased at lower pH.
Figure 2. 2a: vlhA gene copy numbers of 33°C and 39.5°C cultures of MS-H and 86079/7NS determined by vlhA Q-PCR at 24-h intervals. 2b: 33/39.5°C vlhA gene copy number ratios calculated at 24-h intervals. 2c: vlhA gene copy numbers plotted against pH at each time point. 2d: 33/39.5°C vlhA gene copy ratios plotted against pH at each time point. 2e: The vlhA gene copy number of MS-H cultures at 33°C and 39.5°C harvested at different pH irrespective of the time. 2f: 33/39.5°C vlhA gene copy number ratio of MS-H cultures at 33°C and 39.5°C harvested at different pH.
Mathematical calculation to determine the ts phenotype by vlhA quantitative real-time PCR
To examine the capacity of vlhA Q-PCR as a tool for determination of the ts phenotype of an unknown M. synoviae strain, 23 M. synoviae strains with predetermined ts phenotype by conventional microtitration assay (see above) were tested by vlhA Q-PCR. The MS-H vaccine and its parent strain 86079/7NS were included to serve as ts + and ts − controls, respectively. The vlhA copy number of the cultures ranged from 4.9×101 to 4.43×1010 at 33°C, and from 1.51×102 to 5.86×109 at 39.5°C. The vlhA copy number of ts + cultures ranged from 1.62×106 to 4.43×1010 at 33°C, and from 2.63×103 to 1.52×108 at 39.5°C. For ts − cultures, the vlhA copy numbers ranged from 4.9×101 to 3.28×108 at 33°C, and from 1.51×102 to 5.86×109 at 39.5°C. Using 33/39.5°C vlhA gene copy number ratios of the ts − strains, a cut-off value was generated. The mean 33/39.5°C vlhA gene copy number ratio of 13 strains confirmed as ts − by conventional microtitration method was 0.5 with a standard deviation of 0.7. A value of 2.1 (3×standard deviation) was added to the mean and a cut-off value of 2.6 was established. Any unknown M. synoviae strain/isolate with a 33/39.5°C vlhA gene copy number ratio ≤ 2.6 would thus be characterized as ts −. Using this cut-off point, all 10 M. synoviae strains/isolates that had been found to be ts + by conventional microtitration were classified as ts + and all 13 M. synoviae strains/isolates that had been found to be ts − by conventional microtitration were also classified as ts −. The 33/39.5°C vlhA gene copy number ratios ranged from 0.01 to 2.55 for ts − strains/isolates and from 11 to 4811 for ts + strains/isolates ().
Discussion
Microtitration of M. synoviae cultures followed by incubation at two different temperatures is primarily used to determine the temperature-sensitive phenotype of M. synoviae isolates in MS-H vaccinated flocks (differentiation of MS-H from its ts − revertants). This method usually takes 2 to 3 weeks to complete and is relatively expensive. Results of microtitration are also occasionally difficult to interpret when some ts − M. synoviae cultures only partially change the colour of mycoplasma broth at 39.5°C. As the microtitration procedure is not amenable to automation, an attempt was made to develop a rapid molecular technique that can be used in diagnostic laboratories to replace the microtitration procedure.
The quantitative real-time PCR has gained much acceptance in the area of diagnosis and, as equipment and reagents become cheaper and assays become more available, it has become part of routine diagnostic procedures. A real-time Taqman PCR has been developed for detection of M. synoviae (Raviv & Kleven, Citation2009) and shown to be highly sensitive. However, it uses a Taqman probe that is relatively expensive. The Q-PCR developed in this study used a relatively inexpensive intercalating dye and was shown to successfully differentiate ts − and ts + M. synoviae strains/isolates. Specificity of oligonucleotide primers (link-F and MS-Cons-R) used to amplify the conserved region at the 5′ end of the vlhA gene has been described in a previous study (Jeffery et al., Citation2007). Copy numbers as low as 10 per reaction were detected, indicating high sensitivity of the vlhA Q-PCR. The vlhA Q-PCR assay developed in this study was highly reliable and reproducible, with the highest 33/39.5°C vlhA gene copy ratio for the ts − strains/isolates being 2.55 and the lowest 33/39.5°C vlhA gene copy ratio for ts + strains being 11.65, thus providing a relatively wide gap between the ts + and ts − cultures.
Harvesting cultures at an appropriate growth phase was found to be crucial for consistent and reliable determination of 33/39.5°C vlhA gene copy ratios. The MS-H vaccine and its ts + reisolates grew poorly at the non-permissive temperature of 39.5°C but grew to high titres and produced a strong colour change (pH ≤ 6.3) at 33°C. All ts + cultures grew only to approximately pH of 7.3 at 39.5°C, an observation which is in accordance with the previous studies (Markham et al., Citation1998a, Citationb). Contrary to this, most of the ts − field strains and ts − reisolates of MS-H grew almost equally at both permissive and non-permissive temperatures. Such ts − cultures always generated 33/39.5°C ratios that were lower than the calculated cut-off of 2.6 established in this study. It is notable that in this study the ts + cultures never produced a pH change beyond 7.3 at 39.5°C while ts − cultures always did (c,d). Harvesting cultures at approximate pH of 6.8 was selected for Q-PCR studies because M. synoviae grows actively to this pH but may lose viability below it (Whithear, Citation1993). It has been reported that mycoplasmas release nucleases in the culture medium (Razin et al., Citation1964; Pollack & Hoffmann, Citation1982; Paddenberg et al., Citation1998), which may induce cell autolysis (Cowen & Smith, Citation1972) and/or degradation of native DNA in overgrown cultures. This may explain the observation in this study that overgrown MS-H cultures generated relatively low 33/39.5°C vlhA gene copy ratios (d,f). Therefore, it is recommended to harvest cultures at a pH of approximately 6.8 to avoid mischaracterization of unknown cultures.
It was notable that for some isolates the values generated by microtitration versus Q-PCR were disproportional. Most notably, the isolate 94036/8-3a generated a relatively high CCU/ml at both temperatures whilst showing a very low vlhA copy number in Q-PCR at both temperatures. Repeated Q-PCR tests using this isolate did not significantly increase the vlhA copy numbers (results not shown) and it was therefore speculated that this discrepancy could have been caused by primer mismatch. However, for a reason unknown to the authors, several attempts to amplify a larger area of the vlhA gene for the purpose of gaining insight into the Q-PCR primer sites were unsuccessful. Interestingly, attempts to amplify other genes from this isolate as part of an independent study proved very inefficient. Nevertheless, analysis of the amplicon generated using Q-PCR primers revealed that this isolate possessed a vlhA conserved region that was clearly distinct from that of the MS-H (results not shown), reflecting that this isolate was not a “MS-H reisolate” despite being isolated from MS-H vaccinated flocks. It should be noted that the starting cultures used for PCR and microtitration were not necessarily the same, and were occasionally of different passages and/or dilutions. In addition, these two experiments were independently conducted, often at different days or times. Therefore strong correlation between the two results may not always be expected. This may explain disproportional results from these two experiments on the isolate 93198/5-10a, for which a relatively low CCU/ml was obtained, particularly at 39.5°C, while a relatively high vlhA copy number was obtained at the same temperature. Irrespective of the correlation between the results from these two independent experiments, the outcomes (as ts + or ts −) of both experiments were always in agreement.
The revelation of sequence variation between 94036/8-3a and MS-H prompted us to examine the sequence of the Q-PCR amplicons from all other isolates used in our study. Results of this examination confirmed that all had identical vlhA conserved sequence to that of MS-H.
The average time taken by 23 M. synoviae cultures tested in this study to reach the approximate pH 6.8 (either for 33°C or 39.5°C cultures) at 1/5, 1/10 and 1/20 dilution was 43, 50 and 65 h, respectively. Use of multiple dilutions of inoculating culture was found to be a convenient strategy to provide flexibility for monitoring of the growth and with the time of harvest at an appropriate growth phase (pH ~6.8).
This is the first report of a real-time PCR using intercalating dye applied for quantification of M. synoviae. We conclude that vlhA Q-PCR performed on M. synoviae cultures grown at permissive (33°C) and non-permissive (39.5°C) temperatures simultaneously, is a rapid, reliable and cost-effective technique for determination of the ts phenotype of M. synoviae strains. This technique is particularly useful in countries where MS-H vaccine is widely used in commercial poultry and local field strains may be indistinguishable from MS-H vaccine strain by analysis of the vlhA gene.
Supplementary material
Download PDF (649.3 KB)Acknowledgements
The senior author was supported by a scholarship from Higher Education Commission, Pakistan.
References
- Benčina , D. , Drobnič-Valič , M. , Horvat , S. , Narat , M. , Kleven , S.H. and Dovč , P. 2001 . Molecular basis of the length variation in the N-terminal part of Mycoplasma synoviae hemagglutinin . FEMS Microbiology Letters , 203 : 115 – 123 . doi: 10.1111/j.1574-6968.2001.tb10829.x
- Cowen , B.S. and Smith , S.C. 1972 . Nuclease activities of Mycoplasma gallisepticum as a function of culture age in different media . Journal of Bacteriology , 109 : 21 – 24 .
- Feberwee , A. , de Wit , J.J. and Landman , W.J.M. 2009 . Induction of eggshell apex abnormalities by Mycoplasma synoviae: field and experimental studies . Avian Pathology , 38 : 77 – 85 . doi: 10.1080/03079450802662772
- Feberwee , A. and Landman , W.J.M. 2010 . Induction of eggshell apex abnormalities in broiler breeder hens . Avian Pathology , 39 : 133 – 137 . doi: 10.1080/03079451003657637
- Hammond , P.P. , Ramírez , A.S. , Morrow , C.J. and Bradbury , J.M. 2009 . Development and evaluation of an improved diagnostic PCR for Mycoplasma synoviae using primers located in the haemagglutinin encoding gene vlhA and its value for strain typing . Veterinary Microbiology , 136 : 61 – 68 . doi: 10.1016/j.vetmic.2008.10.011
- Harada , K. , Kijima-Tanaka , M. , Uchiyama , M. , Yamamoto , T. , Oishi , K. , Arao , M. and Takahashi , T. 2009 . Molecular typing of Japanese field isolates and live commercial vaccine strain of Mycoplasma synoviae using improved pulsed-field gel electrophoresis and vlhA gene sequencing . Avian Diseases , 53 : 538 – 543 . doi: 10.1637/8934-052309-Reg.1
- Hong , Y. , García , M. , Leiting , V. , Benčina , D. , Dufour-Zavala , L. , Zavala , G. and Kleven , S.H. 2004 . Specific detection and typing of Mycoplasma synoviae strains in poultry with PCR and DNA sequence analysis targeting the hemagglutinin encoding gene vlhA . Avian Diseases , 48 : 606 – 616 . doi: 10.1637/7156-011504R
- Jeffery , N. , Gasser , R.B. , Steer , P.A. and Noormohammadi , A.H. 2007 . Classification of Mycoplasma synoviae strains using single-strand conformation polymorphism and high-resolution melting-curve analysis of the vIhA gene single-copy region . Microbiology , 153 : 2679 – 2688 . doi: 10.1099/mic.0.2006/005140-0
- Jones , J.F. , Whithear , K.G. , Scott , P.C. and Noormohammadi , A.H. 2006 . Onset of immunity with Mycoplasma synoviae: comparison of the live attenuated vaccine MS-H (Vaxsafe MS) with its wild-type parent strain (86079/7NS) . Avian Diseases , 50 : 82 – 87 . doi: 10.1637/7428-083005R.1
- Kleven , S.H. and Ferguson-Noel , N. 2008 . “ Mycoplasma synoviae infection ” . In Diseases of Poultry , 12th edn , Edited by: Saif , Y.M. , Fadly , A. M. , Glisson , J.R. , McDougald , L.R. , Nolan , L.K. and Swayne , D.E. 845 – 857 . Ames , Iowa : Blackwell .
- Markham , J.F. , Morrow , C.J. , Scott , P.C. and Whithear , K.G. 1998a . Safety of a temperature-sensitive clone of Mycoplasma synoviae as a live vaccine . Avian Diseases , 42 : 677 – 681 . doi: 10.2307/1592702
- Markham , J.F. , Scott , P.C. and Whithear , K.G. 1998b . Field evaluation of the safety and efficacy of a temperature-sensitive Mycoplasma synoviae live vaccine . Avian Diseases , 42 : 682 – 689 . doi: 10.2307/1592703
- Meynell , G.G. and Meynell , E. 1970 . Theory and Practice in Experimental Bacteriology , 232 – 233 . Cambridge : Cambridge University Press .
- Morrow , C.J. , Markham , J.F. and Whithear , K.G. 1998 . Production of temperature-sensitive clones of Mycoplasma synoviae for evaluation as live vaccines . Avian Diseases , 42 : 667 – 670 . doi: 10.2307/1592700
- Noormohammadi , A.H. , Jones , J.F. , Harrigan , K.E. and Whithear , K.G. 2003 . Evaluation of the non-temperature-sensitive field clonal isolates of the Mycoplasma synoviae vaccine strain MS-H . Avian Diseases , 47 : 355 – 360 . doi: 10.1637/0005-2086(2003)047[0355:EOTNFC]2.0.CO;2
- Noormohammadi , A.H. , Markham , P.F. , Kanci , A. , Whithear , K.G. and Browning , G.F. 2000 . A novel mechanism for control of antigenic variation in the haemagglutinin gene family of Mycoplasma synoviae . Molecular Microbiology , 35 : 911 – 923 . doi: 10.1046/j.1365-2958.2000.01766.x
- Ogino , S. , Munakata , Y. , Ohashi , S. , Fukui , M. , Sakamoto , H. , Sekiya , Y. , Noormohammadi , A.H. and Morrow , C.J. 2011 . Genotyping of Japanese field isolates of Mycoplasma synoviae and rapid molecular differentiation from the MS-H vaccine strain . Avian Diseases , 55 : 187 – 194 . doi: 10.1637/9461-071310-Reg.1
- Olson , N.O. 1956 . Studies of infectious synovitis in chickens . The American Journal of Veterinary Research , 17 : 747 – 754 .
- Paddenberg , R. , Weber , A. , Wulf , S. and Mannherz , H.G. 1998 . Mycoplasma nucleases able to induce internucleosomal DNA degradation in cultured cells possess many characteristics of eukaryotic apoptotic nucleases . Cell Death and Differentiation , 5 : 517 – 528 . doi: 10.1038/sj.cdd.4400380
- Pollack , J.D. and Hoffmann , P.J. 1982 . Properties of the nucleases of mollicutes . Journal of Bacteriology , 152 : 538 – 541 .
- Ramírez , A.S. , Naylor , C.J. , Yavari , C.A. , Dare , C.M. and Bradbury , J.M. 2011 . Analysis of the 16S to 23S rRNA intergenic spacer region of Mycoplasma synoviae field strains . Avian Pathology , 40 : 79 – 86 . doi: 10.1080/03079457.2010.537305
- Raviv , Z. and Kleven , S.H. 2009 . The development of diagnostic real-time TaqMan PCRs for the four pathogenic avian mycoplasmas . Avian Diseases , 53 : 103 – 107 . doi: 10.1637/8469-091508-Reg.1
- Razin , S. , Knyszynski , A. and Lifshitz , Y. 1964 . Nucleases of mycoplasma . Journal of General Microbiology , 36 : 323 – 332 . doi: 10.1099/00221287-36-2-323
- Whithear , K.G. 1993 . “ Avian mycoplasmosis ” . In Australian Standard Diagnostic Techniques for Animal Diseases , Edited by: Corner , L.A. and Bagust , T.J. 59 – 63 . East Melbourne , , Australia : CSIRO for the Standing Committee on Agriculture and Resource Management .