Abstract
Infectious laryngotracheitis virus (ILTV) causes severe respiratory disease in poultry throughout the world. Recently the role of glycoprotein G (gG) in ILTV pathogenesis has been investigated and it has been shown to have chemokine-binding activity. An ILTV vaccine candidate deficient in gG has been developed and the deletion has been shown to alter the host's immune response to the virus. To understand the effect of the gG gene on transcription of other viral genes, the global expression profile of 72 ILTV genes in gG-deleted and wild-type ILTVs were investigated both in vivo and in vitro using quantitative reverse transcription-polymerase chain reaction. Several genes were differentially expressed in the different viruses in LMH cell cultures or in the tracheas of infected birds, and the expression of a number of genes, including ICP27, gC, gJ, Ul7 and UL40, differed significantly both in vivo and in vitro, suggesting that they had direct or indirect roles in virulence. This study has provided insights into the interactions between gG and other ILTV genes that may have a role in virulence.
Introduction
Over the past few years, a number of studies have been conducted to identify virulence genes in infectious laryngotracheitis virus (ILTV). The effects on virulence of several ILTV genes, including those for TK (Han et al., Citation2002), gC (Pavlova et al., 2009), gJ (Fuchs et al., Citation2005b), UL0 (Veits et al., Citation2003a), UL47 (Helferich et al., Citation2007b), UL50 (Fuchs et al., Citation2000) and open reading frames A to E (Veits et al., Citation2003b), have been investigated by deletion of the corresponding gene and comparison in vitro, and in some cases in vivo, with the parental wild-type strain.
Such studies have identified glycoprotein G (gG) as a virulence factor in ILTV, with gG-deleted ILTV shown to be less pathogenic than the currently available Australian commercial vaccine strains SA-2 and A20 (Devlin et al., Citation2006; Devlin et al., Citation2007). Birds inoculated with gG-deleted ILTV were protected against clinical signs of disease following challenge with virulent ILTV, suggesting that gG-deleted ILTV could have potential as a vaccine (Devlin et al., Citation2007; Coppo et al., Citation2011). In another study, an ILTV mutant in which both the gG and UL47 genes were deleted grew to lower titres in cell culture (Helferich et al., Citation2007a). The gGs of several alphaherpesviruses—including equine herpesvirus-1 and equine herpesvirus-3, bovine herpesvirus-1 and bovine herpesvirus-5, and caprine herpesvirus-1—have been shown to act as viral chemokine-binding proteins in vitro (Bryant et al., Citation2003).
The critical role of chemokines in antiviral defence has been highlighted by the discovery that both poxviruses and herpesviruses encode proteins that mimic chemokines or chemokine receptors and secrete chemokine-binding proteins, which competitively bind and inhibit the interactions of chemokines with cognate receptors. For instance, MT-7 of myxoma virus and M-T1 in vaccinia, myxoma, cowpox and variola viruses bind multiple chemokines with high affinity and inhibit their interaction with chemokine receptors as part of a strategy to prevent the early phase of inflammatory cell migration into virus-infected tissues (Graham et al., Citation1997; Lalani & McFadden, Citation1997).
It has been shown that ILTV gG binds to chemokines with high affinity and inhibits leukocyte chemotaxis (Devlin et al., Citation2010). Chickens infected with gG-deleted ILTV had altered tracheal leukocyte populations and lower serum antibody levels compared with those infected with the parental virus, suggesting that the lack of chemokine-binding activity during infection with gG-deficient ILTV results in altered host immune responses (Devlin et al., Citation2010). To date, the effect of deletion of the gG gene on the expression of other ILTV genes has not been studied. This study thus aimed to gain a better understanding of the effect that gG has on the transcription of other ILTV genes by comparing transcription of wild-type ILTV with that of a gG-deletion mutant in vitro and in vivo using quantitative reverse transcription polymerase chain reaction (qRT-PCR).
Materials and Methods
Viruses and cell culture
The CSW-1 wild-type strain and a gG-deletion mutant derived from it (Devlin et al., Citation2006) were grown in the LMH chicken hepatoma cell line (Kawaguchi et al., Citation1987). LMH cells seeded onto cell culture plates pretreated with 0.1% gelatin (Sigma-Aldrich, Castle Hill, New South Wales, Australia) were incubated at 37°C in 5% carbon dioxide in air in Dulbecco's modified Eagle's medium (Sigma-Aldrich, St. Louis, Missouri, USA) containing 10% foetal bovine serum, 50 mM sodium bicarbonate and 100 µg ampicillin/ml without additional l-glutamine. Cells were infected with 0.1 plaque-forming units of ILTV/cell, and after 30 min (virus adsorption period) excess virus was removed using sterile cold phosphate-buffered saline. The cultures were then incubated at 37°C, and at 12 h post infection (h.p.i.) the media were removed and cells were gently washed with cold phosphate-buffered saline and then used for RNA extraction.
Bird experiments
Ten 3-week-old specific-pathogen-free hybrid White Leghorn chickens (Charles River Laboratories Pty Ltd., Woodend, Victoria, Australia) were weighed and randomly assigned to two groups of five birds and housed in separate negative-pressure fibreglass isolators and provided irradiated feed and water ad libitum. Birds within each group were then inoculated inside the isolator by the intra-tracheal route with a 300 µl dose containing 103 median egg infectious doses of the relevant ILTV strain. Four days after inoculation, birds were examined for clinical signs, weighed and euthanized by exposure to halothane vapour.
RNA extraction
For RNA extraction from LMH cell cultures, the supernatant was removed and the cells washed three times with 2 ml sterile cold phosphate-buffered saline, and then the RNA was extracted using the RNeasy Mini kit (Qiagen, Doncaster, Victoria, Australia) according to the manufacturer's instructions.
For RNA extraction from infected birds, one centimetre of the upper trachea just below the epiglottis was excised and immersed in RNAlater® (Ambion, Austin, Texas, USA), and then stored at −70°C for RNA extraction at a later time. Immediately after thawing, the tracheal samples were processed for RNA extraction by cutting them into small pieces using a scalpel blade, incubating them in 100 µl proteinase K (10 mg/ml) solution (Roche, Mannheim, Germany) at 37°C for 1 h and then preparing a homogeneous mixture in Qiagen lysis buffer (Qiagen) by repeated passage through a three-way tap. The RNA was then extracted using an RNeasy Fibrous Tissue kit (Qiagen) according to the manufacturer's instructions.
The amount of RNA was determined using a Biophotometer spectrophotometer (Eppendorf, Hamburg, Germany).
Preparation of cDNA
Two micrograms of total RNA extracted from tracheal samples or from infected LMH cell cultures was treated with DNase I (Invitrogen, Mulgrave, Victoria, Australia), and cDNA was generated using Superscript™ II RNase H negative SSII reverse transcriptase (Invitrogen) and 100 ng random hexamers (Geneworks, Adelaide, South Australia, Australia). The reactions were terminated by incubation at 70°C for 15 min. A no-RT control that did not contain reverse transcriptase was included for each RNA sample. The total cDNA samples were diluted five-fold with sterile water and aliquots were stored at −20°C.
Quantitative PCR assays
Oligonucleotide primers were manufactured by Geneworks and Invitrogen (Auckland, New Zealand). The sequence and other features of oligonucleotide primers used in this study, including the optimal ratios of each forward and reverse primer that yielded the lowest cycle threshold (Ct) value, have been described previously (Mahmoudian et al., Citation2012). Quantitative PCR was performed using an Mx3000 qPCR thermal cycler (Agilent Technologies, Santa Clara, California, USA). Each PCR reaction (a total volume of 20 µl) contained 10 µl Platinum® SYBR Green qPCR SuperMix-UDG (Invitrogen), 5 µl cDNA and 2.5 µl each primer (0.25 to 0.5 mM). Reactions were incubated for 2 min at 50°C, then 2 min at 95°C, followed by 45 cycles of 60°C for 30 sec and 95°C for 30 sec. Dissociation curve analysis was then performed by ramping the temperature from 55 to 95°C at the instrument default rate of 0.2°C/sec. A reverse transcription negative control (no-RT control, see above) and a negative template control to which no cDNA was added were included in each amplification experiment. Each assay was performed in duplicate and the mean Ct for the duplicate assays used as the value for that gene for that sample for further analyses.
Statistical analysis of expression levels
The Ct values for each sample were normalized against those obtained for two internal control genes—CyP, encoding cyclophilin (GenBank accession number XM_426283) and HPRT-1, encoding hypoxanthine phosphoribosyl-transferase 1 (GenBank accession number NM_204848)—and adjusted using the method of Pfaffl (Citation2001).
The Ct values were further corrected to account for variation in the efficiencies of amplification in each reaction as described previously (Mahmoudian et al., 2012). To determine the level of transcription of each virus, RNA concentrations were analysed in tracheal samples taken from three birds at day 4 after infection and in three independent LMH cell cultures at 12 h.p.i. To compare the level of transcription of each gene in the two strains (as shown in and ), a relative expression level (RelCt) was calculated by subtracting the mean Ct value for all the genes of the strain for the sample from the mean Ct value for each gene for each of the three biological replicate samples:
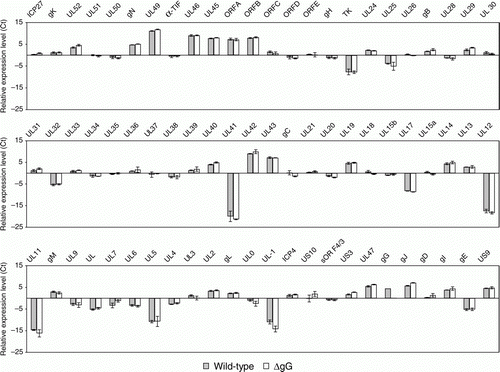
Figure 1. Mean relative levels of transcription of genes of wild-type and gG-deleted ILTV in LMH cells at 12 h.p.i. Error bars indicate the standard deviations for each gene calculated from assays on three independently infected cell cultures.
The mean relative Ct values for the three biological replicates for each gene in the two strains were analysed using Student's t test and P ≤0.05 was considered significant.
Results
Transcription of genes of wild-type and gG-deleted ILTV in LMH cells
The transcription of genes of wild-type and gG-deleted ILTV was investigated at 12 h.p.i. in LMH cells. Genes for which there was a significant difference in mRNA concentrations between the two viruses are listed in and the relative expression levels of all genes are shown in . Apart from the gene for gG, the expression of which was detected only in cells infected with the wild-type strain, significant differences were detected for the genes gC, gE, gD gI, gJ, ICP27, sORF4/3, TK, UL40, UL33, UL6, UL7, UL13 and UL8.
Table 1. Genes with significantly different levels of expression in wild-type ILTV compared with gG-deleted ILTV in either LMH cells and/or the tracheas of infected birds.
Transcription of genes of wild-type and gG-deleted ILTV in the trachea
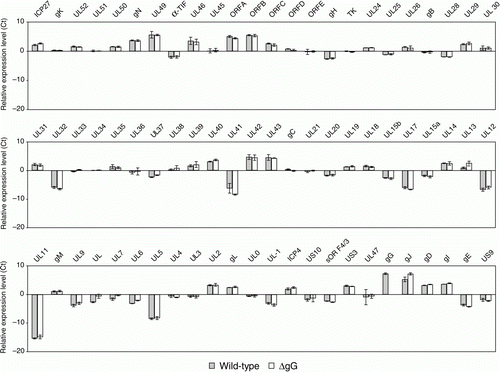
The transcription of genes of wild-type and gG-deleted ILTV in the trachea were determined 4 days after infection of the birds. Genes for which there was a significant difference between the two viruses are listed in and the relative expression levels of all genes are shown in . Apart from the gene for gG, the expression of which was detected only in tracheas of birds infected with the wild-type strain, significant differences were detected for the genes for gC, gJ, gN, ICP27, UL[-1], UL7, UL15b, UL20, UL26, UL29, UL31, UL36, UL39, UL40, UL42, UL47, UL49, UL52 and US3.
Discussion
The tracheal samples were collected 4 days after infection, during the acute phase of disease. It has been shown in previous studies that the concentrations of infectious virus are highest in tracheal washings between 2 and 4 days after infection (Bagust et al., Citation1986), and at this time point there is extensive haemorrhage and cellular infiltration in the tracheal mucosa, which is the major site for ILTV replication. In chickens, it has been shown previously that gG-deleted ILTV reaches a similar titre in the trachea to its wild-type parent, but that its pathogenicity is markedly attenuated, as demonstrated by less severe clinical signs, improved weight gain and enhanced bird survival. Thus, attenuation is not a consequence of reduced viral replication (Devlin et al., Citation2006).
To determine the effect of deletion of gG on the expression of other viral genes, a qRT-PCR assay was used to investigate the concentrations of transcripts from each of the genes of wild-type and gG-deleted ILTV in vivo and in vitro. The analysis of the relative expression levels of the genes of wild-type and gG-deleted ILTV detected significant differences in expression for a number of genes in the trachea or in LMH cell cultures.
Five genes (gC, gJ, ICP27, UL7 and UL40) were differentially expressed both in vivo and in vitro (). Apart from the gC gene, which was more highly expressed in wild-type ILTV, the other genes were more highly expressed in gG-deleted ILTV both in vivo and in vitro.
The gJ gene, which is adjacent to the gG gene, has been found in a separate study to be expressed late in infection and has similar expression kinetics to the gG gene (Mahmoudian et al., Citation2012). The function of ILTV gJ has not yet been fully determined, although in vitro studies have shown that, while deletion of the gJ gene has only minor effects on direct cell-to-cell spread, as measured by plaque size, titres of progeny virus are significantly reduced in comparison with those of the parental virus. It has been shown that gJ is dispensable for replication in chickens (Fuchs et al., Citation2005a), but that it plays an important role during egress of the virus from infected cells in chick embryos (Mundt et al., Citation2011), and that gJ-deleted ILTV is significantly attenuated in chickens (Fuchs et al., 2005b). The gJ homologues in equine herpesvirus-1 and psittacid herpesvirus-1 are dispensable for replication (Sun & Brown, Citation1994; Roizman & Knipe, Citation2001), and herpes simplex virus-1 (HSV-1) gJ has been shown to be a multifunctional protein, modulating several cellular processes, in addition to inhibiting apoptosis (Aubert et al., 2008; Jerome et al., Citation1999). Moreover, it has been shown that HSV-1 gJ inhibits lysis of HSV-1 infected cells by cytotoxic T lymphocytes (Aubert et al., Citation2006). While a similar role for ILTV gJ in immune evasion remains to be determined, its contribution to the virulence of gG-deleted ILTV could be investigated by generating a gG/gJ double knockout mutant.
The higher levels of expression of the gJ gene in gG-deleted ILTV could indicate a suppressive effect of gG on the expression of the gJ gene, compensatory expression in the absence of the immune suppressive effects of gG, or, more likely, that deletion may have had a local effect on the promoter of the gJ gene.
The expression of genomically distant genes, including ICP27, UL7, UL40 and gC, both in vitro and in vivo, was also affected by the gG deletion. Our prior studies of the kinetics of expression of these genes has shown that these genes have early kinetics of expression (Mahmoudian et al., Citation2012), suggesting a role in viral DNA replication or regulation of cellular machinery in favour of viral replication.
The ICP27 gene in HSV-1 is an essential RNA-binding protein that shuttles between the nucleus and cytoplasm to increase cytoplasmic accumulation of late mRNAs and regulates multiple steps of mRNA synthesis and processing, including transcription, splicing and nuclear export (Mears & Rice, Citation1998; Koffa et al., Citation2001). ICP27 is able to induce HSV-1 specific cytotoxic T lymphocytes (Banks et al., Citation1991), and type I interferon-induced STAT-1 nuclear accumulation to avoid the innate immune response (Johnson et al., Citation2008). It has also been shown that the anti-apoptotic effects of ICP27 and ICP22 in HSV-1 are mediated in part through effects on gJ expression (Aubert et al., Citation2008). A similar relationship between the gJ and ICP27 genes in ILTV remains unknown and needs to be investigated. The higher level of expression of ICP27 gene in gG-deleted ILTV may suggest a synergistic role in immune evasion for ICP27 and gG or a suppressive effect of gG on ICP27.
The UL40 product is the small subunit of ribonucleotide reductase in HSV-1, which controls the concentration of deoxyribonucleotides (Roizman & Knipe, Citation2001), The leader sequence of the non-homologue UL40 gene in HCMV can up-regulate expression of HLA-E, which helps infected cells to evade the host immune response by protecting them from NK cell recognition (Cerboni et al., Citation2001). No known interactions with the immune response or similarity in the sequence of UL40 in alphaherpesviruses have been shown. Thus it is hard to determine whether UL40 in ILTV can influence the immune responses or there are other possibilities that UL40 might be differentially expressed.
The UL7 gene was about five-fold and three-fold more highly expressed in gG-deleted ILTV than the parental virus in the tracheas and the LMH cell cultures, respectively (). Homologues of the UL7 gene of HSV-1 are conserved in alphaherpesviruses, betaherpesviruses and gammaherpesviruses. UL7-deleted HSV-1 is replication competent, but forms smaller plaques and yields up to 100-fold fewer progeny than the wild-type virus in Vero cells, and it has been suggested that UL7 may play a supplementary role in DNA cleavage and packaging (Nozawa et al., Citation2002). The UL7 gene of pseudorabies virus has been shown to encode a non-essential structural protein that is involved in virion formation and egress (Fuchs et al., 2005a). One could suggest that removal of the suppressive effect of the gG gene on the expression of the UL7 gene might result in an increase in UL7 functions, such as DNA processing and/or virion maturation, in gG-deleted ILTV. However, no significant difference has been found in the titre between the two viruses after infection.
Earlier, in separate studies in our laboratory, the gC gene was shown to exhibit early kinetics of expression (Mahmoudian et al., Citation2012). Glycoprotein C has been shown to play a primary role in attachment of HSV-1 to the cell surface (Campadelli-Fiume et al., Citation1991). In HSV-1 gC also mediates immune evasion by affecting multiple aspects of innate and acquired immunity, including interfering with the complement components C1q, C3, C5 and properdin, and blocking antibody-dependent cellular cytotoxicity (Lubinski et al., Citation2002). Although complement-binding activity has not yet been demonstrated for ILTV gC, it has been suggested that gC may play a role in immune evasion and it has been shown that gC-deleted ILTV is attenuated (Pavlova et al., 2009).
The expression of a second group of genes was affected only inside the body by deletion of the gG gene (). The absence of an immune response in LMH cell cultures could influence differential expression on in vivo and in vitro. The genes that had higher levels of expression in vivo in the gG-deleted virus could be important for virus survival inside the body in the absence of gG, and their products may complement the functions of gG and assist ILTV in combating the host immune system. For instance, in HSV-1 US3 inhibits cytotoxic T lymphocytes (Aubert et al., Citation2006). It might be that the US3 of ILTV has a similar immunomodulatory function.
The UL[-1] gene showed the greatest difference in expression between the two viruses in vivo, but a difference was not detected in vitro. It has been suggested that the UL[-1] gene may be involved in DNA cleavage and encapsidation in ILTV and may have a role in modulating host cell gene transcription and in determining host tropism and viral pathogenicity (Ziemann et al., Citation1998). Thus, the higher level of expression of UL[-1] in the parental wild-type ILTV may contribute to its greater pathogenicity.
There are several other genes that were more highly expressed by gG-deleted ILTV in the trachea, including UL15b, gN, UL20, UL39, ICP27, UL36, UL49, UL31, UL47, UL42, UL40, US3, UL29, UL52, gJ and UL7. Apart from UL15b, which is an early/late gene, and gJ, gN, UL36 and UL52, which are late genes, these genes are expressed early in infection (Mahmoudian et al., Citation2012). The function(s) of the homologues of these genes in HSV-1 have been shown to be related to DNA replication and/or DNA packaging. The higher levels of expression of these genes may suggest a higher replication rate of gG-deleted ILTV DNA in the trachea, but no significant differences have been seen in the diameters of plaques or the growth kinetics of gG-deleted ILTV and its wild-type parent in vitro. In addition, gG-deleted and wild-type ILTV have been shown to reach a similar titre in the tracheal mucosa of infected birds (Devlin et al., Citation2006). Thus the higher level of expression of these genes with a role in DNA replication does not appear to result in enhanced viral replication.
A third group of genes had differing levels of expression only in vitro (). The functions of these genes have been investigated in several other herpesviruses. An Fc receptor encoded by HSV-1 consists of gE and gI and two protein kinases, UL13 and US3 (Ng et al., Citation1998). UL13 physically associates with gE and mediates the phosphorylation of gE and gI. The genes for UL13, gE and gI were found to be differentially expressed in LMH cells. The serine/threonine protein kinase, UL13, and gI genes were more highly expressed by gG-deleted ILTV, and the gE gene was more highly expressed by the wild-type parent strain in LMH cell cultures. It is not clear why these genes are not more highly expressed in the trachea, but it could be postulated that the immune response may negatively regulate their expression in vivo.
The UL6, UL7, UL8 and UL33 genes were more highly expressed in LMH cell cultures infected with gG-deleted ILTV. These genes have early kinetics of expression in ILTV (Mahmoudian et al., Citation2012). The UL6 protein of HSV-1 has an essential role in virus entry, preassembly and cleavage of DNA (Lamberti & Weller, Citation1996; Newcomb et al., Citation2001), and UL8, which was expressed more highly than other genes in LMH cells infected with the gG-deleted ILTV, encodes a subunit of the helicase–primase complex that is required for viral DNA replication (Chattopadhyay et al., Citation2006). In addition, UL33 plays a role in viral DNA packaging in HSV-1 (Reynolds et al., Citation2000).
The genes that were more highly expressed in LMH cells infected with gG-deleted virus may complement the functions of gG or gG may have an inhibitory effect on the expression of these genes. In contrast, the lower expression of several genes in LMH cells infected with the gG-deleted virus may suggest a role for gG in activating or augmenting the function of these genes. The fact that a number of genes had different levels of expression in the two viruses only in LMH cells suggests that gG may have a role in regulation of viral gene expression, independent of its role in immune evasion.
To further study the regulatory effects of gG in LMH cells, it would be helpful to determine the relationship between mRNA concentrations and their protein products, as mRNA concentrations do not necessarily correlate with their gene product levels.
The only function described for ILTV gG to date is its chemokine-binding capability. However, the data reported here suggest that gG may play a regulatory role early in infection and may have additional functions that are yet to be investigated. Such additional functions may also have direct or indirect effects on cellular and/or viral proteins, and consequently affect the transcription of the viral genome.
In conclusion, the results described in this study showed that deletion of gG had a profound effect on the expression of other ILTV genes through mechanisms that probably include local effects on adjacent genes, as well as through interactions between gG and the immune response. These results provide directions for further studies to investigate the effects of the genes that had different patterns of expression on the immunogenicity of ILTV, facilitating improvement of the gG-deleted vaccine or enabling the design of new vaccine strains.
Acknowledgements
The senior author was supported by a scholarship from Urmia University, Iran.
References
- Aubert , M. , Chen , Z. , Lang , R. , Dang , C.H. , Fowler , C. , Sloan , D.D. and Jerome , K. R. 2008 . The antiapoptotic herpes simplex virus glycoprotein J localizes to multiple cellular organelles and induces reactive oxygen species formation . Journal of Virology , 82 : 617 – 629 . doi: 10.1128/JVI.01341-07
- Aubert , M. , Krantz , E.M. and Jerome , K.R. 2006 . Herpes simplex virus genes Us3, Us5, and Us12 differentially regulate cytotoxic T lymphocyte-induced cytotoxicity . Viral Immunology , 19 : 391 – 408 . doi: 10.1089/vim.2006.19.391
- Bagust , T.J. , Calnek , B.W. and Fahey , K.J. 1986 . Gallid-1 herpesvirus infection in the chicken. 3. Reinvestigation of the pathogenesis of infectious laryngotracheitis in acute and early post-acute respiratory disease . Avian Diseases , 30 : 179 – 190 . doi: 10.2307/1590631
- Banks , T.A. , Allen , E.M. , Dasgupta , S. , Sandri-Goldin , R. and Rouse , B.T. 1991 . Herpes simplex virus type 1-specific cytotoxic T lymphocytes recognize immediate-early protein ICP27 . Journal of Virology , 65 : 3185 – 3191 .
- Bryant , N.A. , Davis-Poynter , N. , Vanderplasschen , A. and Alcami , A. 2003 . Glycoprotein G isoforms from some alphaherpesviruses function as broad-spectrum chemokine binding proteins . The EMBO Journal , 22 : 833 – 846 . doi: 10.1093/emboj/cdg092
- Campadelli-Fiume , G. , Farabegoli , F. , Di Gaeta , S. and Roizman , B. 1991 . Origin of unenveloped capsids in the cytoplasm of cells infected with herpes simplex virus 1 . Journal of Virology , 65 : 1589 – 1595 .
- Cerboni , C. , Mousavi-Jazi , M. , Wakiguchi , H. , Carbone , E. , Kärre , K. and Söderström , K. 2001 . Synergistic effect of IFN-gamma and human cytomegalovirus protein UL40 in the HLA-E-dependent protection from NK cell-mediated cytotoxicity . European Journal of Immunology , 31 : 2926 – 2935 . doi: 10.1002/1521-4141(2001010)31:10%3C2926::AID-IMMU2926%3E3.0.CO;2-2
- Chattopadhyay , S. , Chen , Y. and Weller , S.K. 2006 . The two helicases of herpes simplex virus type 1 (HSV-1) . Frontiers in Bioscience , 11 : 2213 – 2223 . doi: 10.2741/1964
- Coppo , M.J. , Noormohammadi , A.H. , Hartley , C.A. , Gilkerson , J.R. , Browning , G.F. and Devlin , J.M. 2011 . Comparative in vivo safety and efficacy of a glycoprotein G-deficient candidate vaccine strain of infectious laryngotracheitis virus delivered via eye drop . Avian Pathology , 40 : 411 – 417 . doi: 10.1080/03079457.2011.588686
- Devlin , J.M. , Browning , G.F. , Hartley , C.A. and Gilkerson , J.R. 2007 . Glycoprotein G deficient infectious laryngotracheitis virus is a candidate attenuated vaccine . Vaccine , 25 : 3561 – 3566 . doi: 10.1016/j.vaccine.2007.01.080
- Devlin , J.M. , Browning , G.F. , Hartley , C.A. , Kirkpatrick , N.C. , Mahmoudian , A. , Noormohammadi , A.H. and Gilkerson , J.R. 2006 . Glycoprotein G is a virulence factor in infectious laryngotracheitis virus . Journal of General Virology , 87 : 2839 – 2847 . doi: 10.1099/vir.0.82194-0
- Devlin , J.M. , Viejo-Borbolla , A. , Browning , G.F. , Noormohammadi , A.H. , Gilkerson , J.R. , Alcami , A. and Hartley , C.A. 2010 . Evaluation of immunological responses to a glycoprotein G deficient candidate vaccine strain of infectious laryngotracheitis virus . Vaccine , 28 : 1325 – 1332 . doi: 10.1016/j.vaccine.2009.11.013
- Fuchs , W. , Granzow , H. , Klopfleisch , R. , Klupp , B.G. , Rosenkranz , D. and Mettenleiter , T.C. 2005a . The UL7 gene of pseudorabies virus encodes a nonessential structural protein which is involved in virion formation and egress . Journal of Virology , 79 : 11291 – 11299 . doi: 10.1128/JVI.79.17.11291-11299.2005
- Fuchs , W. , Wiesner , D. , Veits , J. , Teifke , J.P. and Mettenleiter , T.C. 2005b . In vitro and in vivo relevance of infectious laryngotracheitis virus gJ proteins that are expressed from spliced and nonspliced mRNAs . Journal of Virology , 79 : 705 – 716 . doi: 10.1128/JVI.79.2.705-716.2005
- Fuchs , W. , Ziemann , K. , Teifke , J.P. , Werner , O. and Mettenleiter , T.C. 2000 . The non-essential UL50 gene of avian infectious laryngotracheitis virus encodes a functional dUTPase which is not a virulence factor . Journal of General Virology , 81 : 627 – 638 .
- Graham , K.A. , Lalani , A.S. , Macen , J.L. , Ness , T.L. , Barry , M. , Liu , L.Y. , Lucas , A. , Clark-Lewis , I. , Moyer , R.W. and McFadden , G. 1997 . The T1/35kDa family of poxvirus-secreted proteins bind chemokines and modulate leukocyte influx into virus-infected tissues . Virology , 229 : 12 – 24 . doi: 10.1006/viro.1996.8423
- Han , M.G. , Kweon , C.H. , Mo , I.P. and Kim , S.J. 2002 . Pathogenicity and vaccine efficacy of a thymidine kinase gene deleted infectious laryngotracheitis virus expressing the green fluorescent protein gene . Archives of Virology , 147 : 1017 – 1031 . doi: 10.1007/s00705-001-0794-y
- Helferich , D. , Veits , J. , Mettenleiter , T.C. and Fuchs , W. 2007a . Identification of transcripts and protein products of the UL31, UL37, UL46, UL47, UL48, UL49 and US4 gene homologues of avian infectious laryngotracheitis virus . Journal of General Virology , 88 : 719 – 731 . doi: 10.1099/vir.0.82532-0
- Helferich , D. , Veits , J. , Teifke , J.P. , Mettenleiter , T.C. and Fuchs , W. 2007b . The UL47 gene of avian infectious laryngotracheitis virus is not essential for in vitro replication but is relevant for virulence in chickens . Journal of General Virology , 88 : 732 – 742 . doi: 10.1099/vir.0.82533-0
- Jerome , K.R. , Fox , R. , Chen , Z. , Sears , A.E. , Lee , H. and Corey , L. 1999 . Herpes simplex virus inhibits apoptosis through the action of two genes, Us5 and Us3 . Journal of Virology , 73 : 8950 – 8957 .
- Johnson , K.E. , Song , B. and Knipe , D.M. 2008 . Role for herpes simplex virus 1 ICP27 in the inhibition of type I interferon signaling . Virology , 374 : 487 – 494 . doi: 10.1016/j.virol.2008.01.001
- Kawaguchi , T. , Nomura , K. , Hirayama , Y. and Kitagawa , T. 1987 . Establishment and characterization of a chicken hepatocellular carcinoma cell line, LMH . Cancer Research , 47 : 4460 – 4464 .
- Koffa , M.D. , Clements , J.B. , Izaurralde , E. , Wadd , S. , Wilson , S.A. , Mattaj , I.W. and Kuersten , S. 2001 . Herpes simplex virus ICP27 protein provides viral mRNAs with access to the cellular mRNA export pathway . The EMBO Journal , 20 : 5769 – 5778 . doi: 10.1093/emboj/20.20.5769
- Lalani , A.S. and McFadden , G. 1997 . Secreted poxvirus chemokine binding proteins . Journal of Leukocyte Biology , 62 : 570 – 576 .
- Lamberti , C. and Weller , S.K. 1996 . The herpes simplex virus type 1 UL6 protein is essential for cleavage and packaging but not for genomic inversion . Virology , 226 : 403 – 407 . doi: 10.1006/viro.1996.0668
- Lubinski , J.M. , Jiang , M. , Hook , L. , Chang , Y. , Sarver , C. , Mastellos , D. , Lambris , J.D. , Cohen , G.H. , Eisenberg , R.J. and Friedman , H.M. 2002 . Herpes simplex virus type 1 evades the effects of antibody and complement in vivo . Journal of Virology , 76 : 9232 – 9241 . doi: 10.1128/JVI.76.18.9232-9241.2002
- Mahmoudian , A. , Markham , P.F. , Noormohammadi , A.H. and Browning , G.F. 2012 . Kinetics of transcription of infectious laryngotracheitis virus genes . Comparative Immunology, Microbiology and Infectious Diseases , 35 : 103 – 115 . doi: 10.1016/j.cimid.2011.11.001
- Mears , W.E. and Rice , S.A. 1998 . The herpes simplex virus immediate-early protein ICP27 shuttles between nucleus and cytoplasm . Virology , 242 : 128 – 137 . doi: 10.1006/viro.1997.9006
- Mundt , A. , Mundt , E. , Hogan , R.J. and Garcia , M. 2011 . Glycoprotein J of infectious laryngotracheitis virus is required for efficient egress of infectious virions from cells . Journal of General Virology , 92 : 2586 – 2589 . doi: 10.1099/vir.0.031443-0
- Newcomb , W.W. , Juhas , R.M. , Thomsen , D.R. , Homa , F.L. , Burch , A.D. , Weller , S.K. and Brown , J.C. 2001 . The UL6 gene product forms the portal for entry of DNA into the herpes simplex virus capsid . Journal of Virology , 75 : 10923 – 10932 . doi: 10.1128/JVI.75.22.10923-10932.2001
- Ng , T.I. , Ogle , W.O. and Roizman , B. 1998 . UL13 protein kinase of herpes simplex virus 1 complexes with glycoprotein E and mediates the phosphorylation of the viral Fc receptor: glycoproteins E and I . Virology , 241 : 37 – 48 . doi: 10.1006/viro.1997.8963
- Nozawa , N. , Daikoku , T. , Yamauchi , Y. , Takakuwa , H. , Goshima , F. , Yoshikawa , T. and Nishiyama , Y. 2002 . Identification and characterization of the UL7 gene product of herpes simplex virus type 2 . Virus Genes , 24 : 257 – 266 . doi: 10.1023/A:1015332716927
- Pavlova , S.P. , Veits , J. , Blohm , U. , Maresch , C. , Mettenleiter , T.C. and Fuchs , W. 2010 . In vitro and in vivo characterization of glycoprotein C-deleted infectious laryngotracheitis virus . Journal of General Virology , 91 : 847 – 857 . doi: 10.1099/vir.0.016634-0
- Pfaffl , M.W. 2001 . A new mathematical model for relative quantification in real-time RT-PCR . Nucleic Acids Research , 29 : e45 doi: 10.1093/nar/29.9.e45
- Reynolds , A.E. , Fan , Y. and Baines , J.D. 2000 . Characterization of the U(L)33 gene product of herpes simplex virus 1 . Virology , 266 : 310 – 318 . doi: 10.1006/viro.1999.0090
- Roizman , B. and Knipe , D.M. 2001 . “ Herpes simplex viruses and their replication ” . In Field's Virology , 4th edn , Edited by: Knipe , D.M. , Howley , P.M. , Griffin , D.E. , Lamb , R.A. , Martin , M.A. , Roizman , B. and Straus , S.E. 2399 – 2459 . Philadelphia : Lippincott Williams & Wilkins .
- Sun , Y. and Brown , S.M. 1994 . The open reading frames 1, 2, 71, and 75 are nonessential for the replication of equine herpesvirus type 1 in vitro . Virology , 199 : 448 – 452 . doi: 10.1006/viro.1994.1143
- Veits , J. , Luschow , D. , Kindermann , K. , Werner , O. , Teifke , J.P. , Mettenleiter , T.C. and Fuchs , W. 2003a . Deletion of the non-essential UL0 gene of infectious laryngotracheitis (ILT) virus leads to attenuation in chickens, and UL0 mutants expressing influenza virus haemagglutinin (H7) protect against ILT and fowl plague . Journal of General Virology , 84 : 3343 – 3352 . doi: 10.1099/vir.0.19570-0
- Veits , J. , Mettenleiter , T.C. and Fuchs , W. 2003b . Five unique open reading frames of infectious laryngotracheitis virus are expressed during infection but are dispensable for virus replication in cell culture . Journal of General Virology , 84 : 1415 – 1425 . doi: 10.1099/vir.0.18926-0
- Ziemann , K. , Mettenleiter , T.C. and Fuchs , W. 1998 . Infectious laryngotracheitis herpesvirus expresses a related pair of unique nuclear proteins which are encoded by split genes located at the right end of the UL genome region . Journal of Virology , 72 : 6867 – 6874 .