Abstract
Antimicrobial-resistant Salmonella enterica poses a particular risk to public health, and in particular isolates belonging to clonal lineages such as Salmonella Typhimurium DT104 cause epidemics across species including poultry. In recent years, antimicrobial-resistant S. Typhimurium DT193 and specifically the monophasic S. Typhimurium-like variants of this phage type, serotypes 4,12:i:- and 4,5,12:i:-, have become an increasing risk to public health in Europe and the USA and now account for nearly one-half of human S. Typhimurium infections in the UK. Unlike S. Typhimurium that possesses two forms of flagella which can vary between phase 1 and phase 2 during infection, monophasic variants possess only phase 1 flagella. These monophasic antimicrobial-resistant variants have become a major problem in pig production but human cases have also been associated with poultry consumption and have been found in UK flocks through surveillance schemes since 2010. In this study we determined the ability of antimicrobial-resistant DT193 serotype 4,12:i:- and 4,5,12:i:- isolates from pigs to infect chickens. All isolates were found to colonize the caeca and liver. All but one isolate of serotype 4,5,12:i:- also infected the spleen. Levels of infection and pathology were comparable with those found with the virulent S. Typhimurium isolate 4/74. These findings indicate that both S. Typhimurium DT193 and monophasic variants of this phage type usually associated with pigs are capable of colonizing the chicken. This shows that both S. Typhimurium DT193 and monophasic variants represent a significant and potential emerging threat to poultry production from “spill-over” of these isolates from the pig industry or other sources.
Introduction
Salmonella enterica remains a major cause of human gastrointestinal disease, with poultry a major worldwide source of human infection (Barrow et al., Citation2012). In recent years there has been a significant reduction in poultry, and in particular eggs, as the infection source of S. Enteritidis in the UK through the introduction of vaccination programmes and improvements in biosecurity and hygiene as set out by National Control Plans (O'Brien, Citation2013). However, despite the success in controlling members of Salmonella in eggs and reduction of salmonella isolations from UK broiler flocks, poultry remain an important source of infection.
In recent years the emergence of antimicrobial-resistant isolates of Salmonella has been an increasing problem in animal production and public health. Among these antibiotic-resistant Salmonella strains perhaps the greatest threat to public health are atypical monophasic variants of S. Typhimurium (EFSA, Citation2010). These are frequently associated with invasive human gastroenteritis (Trupschuch et al., Citation2010; Wasyl & Hoszowski, Citation2012). Unlike conventional S. Typhimurium that utilizes two distinct and variable forms of flagella, termed phase 1 and phase 2 flagella, for extracellular and intracellular invasion and movement, monophasic isolates do not convert to phase 2 (Soyer et al., Citation2009). These “epidemic” monophasic isolates have been found to lack fljB, a gene that encodes phase 2 flagella (Soyer et al., Citation2009; Huehn et al., Citation2010). Monophasic members of Salmonella predominantly belong to two main phage types, DT193 and DT104, and pigs are considered the predominant source (Hauser et al., Citation2010; Huehn et al., Citation2010; Wasyl & Hoszowski, Citation2012). In Germany around 40% of human S. Typhimurium isolates submitted to the National Reference Centre are monophasic variants (Trupschuch et al., Citation2010). Molecular analysis has found that monophasic members of Salmonella isolated from pigs, pork and humans display high levels of genetic homology (Hauser et al., Citation2010). In the UK, DT193 has emerged as a major phage type in human infection with some 48% of human isolates in 2010 of the phage type, and around one-half of these monophasic variants (Hopkins et al., Citation2012), and are also increasingly found in meat and poultry products in the USA (USDA, Citation2012). Monophasic Salmonella serotypes are considered an emerging problem in poultry that has also been implicated as a source of human infection (Huehn et al., Citation2009; DEFRA, Citation2012). The two major DT193 monophasic variants serotype 4,12:i:- and serotype 4,5,12:i:- have recently been isolated for the first time from UK poultry farms, with 12 incidents in 2010 and 2011 indicating their emerging threat to poultry production (DEFRA, Citation2012).
In this study we assess the ability of both S. Typhimurium DT193 and the monophasic variants serotypes 4,12:i:- and 4,5,12:i:- to infect and colonize the chicken. Understanding the ability of monophasic variants to colonize the chicken and hence enter the food chain is key to understanding the emerging public health risk of spread of monophasic S. Typhimurium-like variants from pig to poultry production.
Materials and Methods
Bacterial isolates
The source of the bacterial isolates and their antimicrobial resistance profile is summarized in . S. Typhimurium 4/74 was included as a control isolate of defined virulence and ability to colonize the caeca of chickens (Richardson et al., Citation2011). All isolates were maintained as frozen stock at 80°C on cryoprotective beads and grown as required. Bacteria were grown Luria-Bertani (Oxoid, Basingstoke, UK) broth for 18 h at 37°C in a shaking incubator (150 r.p.m.).
Table 1. Salmonella isolates used in this study and their antimicrobial resistance profile.
In vivo infection experiments
All work was conducted in accordance with UK legislation governing experimental animals under project licences PPL 40/3063 and PPL40/3652 and was approved by the University of Liverpool ethical review process prior to the award of the licence. All birds were checked a minimum of twice daily to ensure their health and welfare. Birds were housed in accommodation meeting UK legislation requirements.
Specified pathogen free (SPF) White Leghorn eggs were obtained from Charles River Laboratories International Inc. (Isaszeg, Hungary) and hatched under high biosecurity conditions in the experimental poultry unit at the University of Liverpool. Chicks were maintained in this unit for work with containment level 2 pathogens within lobbied rooms with individual filtered ventilation and air extraction, and in rooms under negative pressure to prevent pathogen transmission from experimental rooms. Entry into the unit is strictly regulated and all personnel wear dedicated protective clothing in the facility with a separate change of boots and dedicated overalls between each experimental room. Such high levels of biosecurity and a standard operating procedure for work in the unit for research and support staff mean that transmission into, from and between experimental groups is prevented. At 1 day of age, chicks were housed separately in five groups in floor pens at a temperature of 30°C and were given ad libitum access to water and a laboratory-grade vegetable protein-based pellet diet (SDS, Witham, UK). At 8 days of age chicks were inoculated by oral gavage with 108 colony-forming units of the salmonellas. At 3 and 7 days post infection, five birds from each group were killed for post-mortem analysis. The liver, spleen and then the caecal contents were removed aseptically from each bird and diluted 1:10 (wt/vol.) in sterile phosphate-buffered saline. Tissue and caecal content samples were then homogenized in a Colworth 80 microstomacher (A.J. Seward & Co. Ltd, London, UK). Samples were serially diluted and dispensed onto Brilliant Green agar (Oxoid, Cambridge, UK) to quantify numbers of salmonellas as described previously (Salisbury et al., Citation2011).
Statistical analysis
Statistical analysis was performed using SPSS v.20 (IBM, Armonk, NY, USA). Comparison of bacterial load between infected groups through the Kruskal–Wallis test, an equivalent non-parametric test to analysis of variance, was used because the data were not distributed normally.
Results and Discussion
All isolates including both biphasic DT193 and the monophasic variants colonized the gastrointestinal tract (), although isolate S04327-09 (serotype 4,5,12:i) colonized to a significantly lower level than 4/74 at either 3 days post infection (P = 0.016) or 7 days post infection (P = 0.026). In contrast, the other monophasic and biphasic DT193 all colonized to similar levels as 4/74. All isolates were detected in similar numbers of the liver at both 3 and 7 days post infection (b), although isolate S04327-09 (serotype 4,5,12:i-) was not found in the spleen (c). In all infected birds there was mild hepatosplenomegaly at both post-mortem time points and mild hyperaemia in the ileum at 3 days post infection consistent with an inflammatory response in the gut. These findings are consistent with previous infection studies with S. Typhimurium in SPF chickens (Withanage et al., Citation2005). Taken as a whole these finding show that both biphasic DT193 and the monophasic variants serotypes 4,12:i:- and 4,5,12:i:- can colonize the intestines of chickens to a high level and are able to invade systemic sites at a similar level to the well-defined virulent isolate 4/74. Whilst as yet chicken has not been considered the main source of these variants in human infection, it is clear from the data presented here and UK surveillance data that monophasic S. Typhimurium-like serotypes 4,12:i:- and 4,5,12:i:- are a risk to poultry production (Hopkins et al., Citation2012).
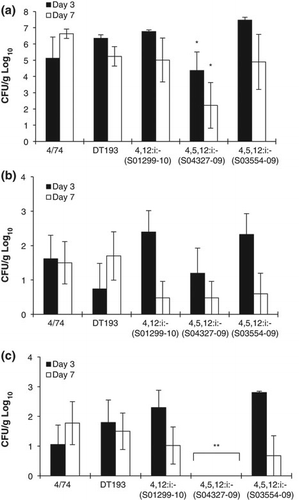
Although largely considered porcine in origin, both DT193 and its monophasic variants can infect and colonize the chicken. Given the high levels of salmonellas in intensive pig production in Europe and the USA—for instance, around one-quarter of pig carcasses in the UK are positive for members of Salmonella at slaughter—there is a considerable threat of spill-over from pig production to other livestock. There were 60 incidents of monophasic S. Typhimurium in pig production premises in the UK in 2011; 38 of these were DT193 isolates, clearly illustrating the extent of this epidemic in pig production. Indeed it is clear that there can be considerable “leakage” of monophasic multi drug-resistant members of Salmonella from pig farms to the environment (Antunes et al., Citation2011). As such the risk of contamination of poultry premises from pig production, and in particular on mixed farms, is potentially significant particularly if there is any breakdown in biosecurity. To some extent there are parallels to the S. Typhimurium DT104 epidemic in the 1990s, in that there is cross-species spread of an antimicrobial-resistant variant that is fully virulent (Threlfall, Citation2000). All of the isolates in this study display a full range of virulence-associated genes and normal growth characteristics (Crayford, Humphrey, & Wigley, unpublished data), suggesting they are capable of causing gastrointestinal disease in humans. This differs from serotype 4,12:d, a monophasic isolate associated with broiler chickens but not considered pathogenic to humans (Huehn et al., Citation2009). It is of some interest that strain S04327-09 was the least invasive and poorest colonizer of the chicken. Its relatively poor infectivity may be a consequence of reduced fitness due to its wide-ranging antimicrobial resistance in comparison with the other monophasic isolates (Table 1), although the biphasic DT193 has a similar profile.
The data presented here provide a clear demonstration that monophasic S. Typhimurium-like isolates of serotypes 4,12:i:- and 4,5,12:i:- may infect the chicken and colonize the gastrointestinal tract, and as such pose a risk to broiler production. Furthermore, although the more stringent biosecurity employed by the poultry industry as part of National Control Plans are likely to mean the risk of transmission to poultry is lower than in pig production, it is clear that entry into poultry production has already occurred and that poultry has been implicated as a source of human infection (Switt et al., Citation2009; DEFRA, Citation2012). Moreover, the efficacy of vaccination against monophasic variants is not yet known, but may differ given the changes to the flagellar antigenic profile and may result in reduced protection in broiler breeder flocks.
Acknowledgements
The authors wish to thank Professors Rob Davies (AHVLA) and Mark Stevens (Roslin Institute, University of Edinburgh) for the isolates used in this study. They also thank Dr Kannan Ganapathy, University of Liverpool for the kind gift of the SPF chicks used in this study. The authors wish to acknowledge the financial support of the Houghton Trust and BPEX for supporting GC's studentship.
References
- Antunes, P., Mourao, J., Pestana, N. & Peixe, L. (2011). Leakage of emerging clinically relevant multidrug-resistant Salmonella clones from pig farms. Journal of Antimicrobial Chemotherapy, 66, 2028–2032.
- Barrow, P.A., Jones, M.A., Smith, A.L. & Wigley, P. (2012). The long view: Salmonella – the last forty years. Avian Pathology, 41, 413–420.
- DEFRA. (2012). Salmonella in Livestock Production in GB. London: DEFRA.
- EFSA. (2010). Scientific opinion on monitoring and assessment of the public health risk of “Salmonella Typhimurium-like” strains. EFSA Journal, 8, 1826.
- Hauser, E., Tietze, E., Helmuth, R., Junker, E., Blank, K., Prager, R., Rabsch, W., Appel, B., Fruth, A. & Malorny, B. (2010). Pork contaminated with Salmonella enterica serovar 4,[5],12:i:-, an emerging health risk for humans. Applied and Environmental Microbiology, 76, 4601–4610.
- Hopkins, K.L., de Pinna, E. & Wain, J. (2012). Prevalence of Salmonella enterica serovar 4,[5],12:i:- in England and Wales, 2010. Euro Surveillance, 17.
- Huehn, S., Bunge, C., Junker, E., Helmuth, R. & Malorny, B. (2009). Poultry-associated Salmonella enterica subsp. enterica serovar 4,12:d:- reveals high clonality and a distinct pathogenicity gene repertoire. Applied and Environmental Microbiology, 75, 1011–1020.
- Huehn, S., La Ragione, R.M., Anjum, M., Saunders, M., Woodward, M.J., Bunge, C., Helmuth, R., Hauser, E., Guerra, B., Beutlich, J., Brisabois, A., Peters, T., Svensson, L., Madajczak, G., Litrup, E., Imre, A., Herrera-Leon, S., Mevius, D., Newell, D.G. & Malorny, B. (2010). Virulotyping and antimicrobial resistance typing of Salmonella enterica serovars relevant to human health in Europe. Foodborne Pathogens and Disease, 7, 523–535.
- O'Brien, S.J. (2013). The “decline and fall” of nontyphoidal salmonella in the United Kingdom. Clinical Infectious Diseases, 56, 705–710.
- Richardson, E.J., Limaye, B., Inamdar, H., Datta, A., Manjari, K.S., Pullinger, G.D., Thomson, N.R., Joshi, R.R., Watson, M. & Stevens, M.P. (2011). Genome sequences of Salmonella enterica serovar Typhimurium, Choleraesuis, Dublin, and Gallinarum strains of well- defined virulence in food-producing animals. Journal of Bacteriology, 193, 3162–3163.
- Salisbury, A.M., Bronowski, C. & Wigley, P. (2011). Salmonella Virchow isolates from human and avian origins in England – molecular characterization and infection of epithelial cells and poultry. Journal of Applied Microbiology, 111, 1505–1514.
- Soyer, Y., Moreno Switt, A., Davis, M.A., Maurer, J., McDonough, P.L., Schoonmaker-Bopp, D.J., Dumas, N.B., Root, T., Warnick, L.D., Grohn, Y.T. & Wiedmann, M. (2009). Salmonella enterica serotype 4,5,12:i:-, an emerging Salmonella serotype that represents multiple distinct clones. Journal of Clinical Microbiology, 47, 3546–3556.
- Switt, A.I.M., Soyer, Y., Warnick, L.D. & Wiedmann, M. (2009). Emergence, distribution, and molecular and phenotypic characteristics of Salmonella enterica serotype 4,5,12:i:-. Foodborne Pathogens and Disease, 6, 407–415.
- Threlfall, E.J. (2000). Epidemic Salmonella typhimurium DT 104-a truly international multiresistant clone. Journal of Antimicrobial Chemotherapy, 46, 7–10.
- Trupschuch, S., Laverde Gomez, J.A., Ediberidze, I., Flieger, A. & Rabsch, W. (2010). Characterisation of multidrug-resistant Salmonella Typhimurium 4,[5],12:i:- DT193 strains carrying a novel genomic island adjacent to the thrW tRNA locus. International Journal of Medical Microbiology, 300, 279–288.
- USDA. (2012). Serotypes profile of Salmonella isolates from meat and poultry products from January 1999 through December 2011. United States Department of Agriculture Report.
- Wasyl, D. & Hoszowski, A. (2012). Occurrence and characterization of monophasic Salmonella enterica serovar Typhimurium (1,4,[5],12:i:-) of non-human origin in Poland. Foodborne Pathogens and Disease, 9, 1037–1043.
- Withanage, G.S., Wigley, P., Kaiser, P., Mastroeni, P., Brooks, H., Powers, C., Beal, R., Barrow, P., Maskell, D. & McConnell, I. (2005). Cytokine and chemokine responses associated with clearance of a primary Salmonella enterica serovar Typhimurium infection in the chicken and in protective immunity to rechallenge. Infection and Immunity, 73, 5173–5182.