Abstract
Staphylococcus aureus is a highly versatile pathogen in a large number of domestic animals, including avian species. To gain deeper insight into the epidemiology and diversity of S. aureus associated with articular disease in domestic turkeys, isolates were collected from infected foot joints of turkeys in Brittany (France). A total of 34 isolates were recovered and characterized by means of antimicrobial resistance, staphylococcal protein A typing, macrorestriction pulsed-field gel electrophoresis and micro-array analysis. Thirty isolates were identified as clonal complex (CC) 398 and methicillin-susceptible S. aureus (MSSA), one was identified as a methicillin-resistant S. aureus (MRSA) CC398 isolate, and the remaining were also MSSA and belonged to CC5, CC101, and CC121. Eleven different antimicrobial resistance patterns were detected, with most isolates resistant to penicillin and tetracycline. Based on all typing methods used, the 34 isolates could be divided into 22 different strains. Results on selected isolates, genotyped using microarrays, indicated a high homogeneity among pathogenic MSSA isolates from turkeys. Moreover, all isolates, except the unique MRSA isolate, carried specific φAvβ prophage avian-niche-specific genes, demonstrating the versatility of S. aureus to adapt to the specific ecological poultry niche.
Introduction
Staphylococcus aureus has long been recognized as an important highly versatile pathogen in both human and veterinary medicine. In poultry, as well as in many other animal species and humans, S. aureus belongs to the normal skin microflora (Devriese, Citation1980). S. aureus can be involved in diseases such as osteomyelitis, bumble foot, and arthritis in poultry. The pathogen can be isolated from the joints, tendon sheaths and bone of affected animals (Andreasen, Citation2003). Former studies have demonstrated the existence of a limited number of S. aureus genotypes associated with carriage and pathology in poultry, the majority of isolates belonging to clonal complex (CC) 5 (Fitzgerald, Citation2012). CC5 is also one of the most successful human-associated lineages (Lindsay, Citation2010). Recent phylogenetic studies have revealed that the poultry CC5 lineage has only recently diverged from this human lineage (Lowder et al., Citation2009). Another common lineage in poultry is CC398, which has been shown to be present also in other host species including humans, pigs, dairy cows and veal calves, horses and companion animals (Vanderhaeghen et al., Citation2012; Fitzgerald, Citation2012). Recently, whole genome sequencing has proved that humans are the ancestral host of this lineage (Price et al., Citation2012).
Most studies involving poultry have focused on chickens while there is little information on turkeys. Recently, studies focusing on the presence of methicillin-resistant S. aureus (MRSA) carriage among turkeys (Richter et al., Citation2012) and turkey meat products (de Boer et al., Citation2009; Feßler et al., Citation2011a) have been published. However there is little research on S. aureus involved in disease in turkeys, with the exception of osteomyelitis cases (Alfonso & Barnes, Citation2006; Corrand et al., Citation2012). Recently, German results showed that livestock-associated MRSA may represent a larger cause of human infection than estimated previously (Köck et al., Citation2013). Turkeys represent a potential source of livestock-associated MRSA, stressing the need for closed surveillance. To gain insight into the S. aureus strains present among diseased turkeys, in this study we investigated the clonal diversity and the genetic characteristics of S. aureus isolates recovered from infected joints of turkeys.
Materials and Methods
Bacterial isolates
A total of 34 S. aureus isolates were analysed in this study. The isolates were recovered by Chêne Vert Conseil (Loudéac, France) and Labofarm (Loudéac, France) and they were obtained between 2008 and 2012 from diseased breeder turkeys. Turkeys originated from three different hatcheries in the region of Brittany (France) and were subsequently raised at different farms. Each S. aureus isolate was recovered from a different bird. Twenty-three isolates were obtained from subcutaneous abscesses of feet (colloquially known as bumble foot) during the laying period, and the remaining isolates were recovered from young birds with arthritis during the rearing period. Bacteria from infection sites were cultivated on Columbia CNA agar (selective medium for Gram-positive bacteria) and Columbia sheep blood agar (Thermo Scientific, Oxoid, Basingstoke, Hampshire, UK). Suspected S. aureus colonies were identified using the Staphylococcus Latex Kit (Plasmatec Laboratory, Bridport, UK).
Genotyping
Genomic DNA was purified as described previously (Vanderhaeghen et al., Citation2010). MRSA was identified using a triplex polymerase chain reaction (PCR) developed for detection of the staphylococcal specific 16S rDNA gene, the S. aureus specific nuc1 gene, and the methicillin resistance mecA gene (Maes et al., Citation2002). The strains were confirmed as belonging to the CC398 using the sau1-hsdS1 CC398 PCR reaction (Stegger et al., Citation2011). Strains were typed by staphylococcal protein A (spa) typing according to the Ridom StaphType standard protocol (www.ridom.de/staphtype). Some isolates were also subjected to multilocus sequence typing (http://www.mlst.net/). Briefly, amplicons of the spa and multilocus sequence typing genes were purified with the Nucleospin Extract II kit (Macherey-Nagel, Düren, Germany) and then sequenced using the CEQ 8000 Genetic Analysis System (Beckman Coulter, Suarlée, UK) according to the manufacturer's instructions. The Ridom StaphType software package (Ridom GmbH, Münster, Germany) and the S. aureus multilocus sequence typing website (http://saureus.mlst.net/) were used to assign spa and sequence types (STs), respectively. SCCmec typing of MRSA isolates were performed using the multiplex PCRs described by Kondo et al. (Citation2007).
Macrorestriction pulsed-field gel electrophoresis analysis
Whole DNA from each S. aureus isolate was analysed by SmaI and Cfr9I macrorestriction and pulsed-field gel electrophoresis (PFGE) using a CHEF Mapper system (Bio-Rad Laboratories, Hemel Hempstead, UK). Plugs were prepared according to the protocol of Argudín et al. (Citation2010c). Slices of the plugs were subjected to restriction with SmaI (Fermentas GmbH, Aalst, Belgium) or Cfr9I (Fermentas GmbH) following the manufacturer's instructions. The running electrophoresis conditions were 6 V/cm in 0.5× TBE (45 mM Tris, 45 mM boric acid, 1 mM ethylenediamine tetraacetic acid, pH 8) at 14°C, and runs lasted 18 h with switch times from 2.63 to 63.8 sec. PFGE profiles were compared digitally using BioNumerics software (Version 6.6; Applied Maths, Laethem-Saint-Martin, Belgium). Cluster analysis of Dice similarity indices based on the unweighted pair group method with arithmetic averages was applied to generate a dendrogram depicting the relationships among PFGE profiles. S. aureus NCTC 8325 (National Collection of Type Cultures, Salisbury, UK) was included as a control strain for PFGE analysis.
Antimicrobial susceptibility testing
Minimal inhibitory concentrations of 19 antimicrobials (penicillin, cefoxitin, kanamycin, streptomycin, gentamicin, erythromycin, clindamycin, quinupristin/dalfopristin, linezolid, tiamulin, chloramphenicol, rifampicin, ciprofloxacin, fusidic acid, tetracycline, trimethoprim, sulfamethoxazole, vancomycin and mupirocin) were determined using custom veterinary international Sensititre staphylococci plates (EUST; Trek Diagnostics System, East Grinstead, UK) according to the manufacturer's instructions. The interpretation of minimal inhibitory concentration values was according to the epidemiological cut-off values of the European Committee for Antimicrobial Susceptibility Testing (EUCAST) for S. aureus. Data from the EUCAST minimal inhibitory concentration distribution website was last accessed on 6 August 2013 (http://www.eucast.org).
DNA microarray-based typing and detection of resistance and virulence genes
Twelve isolates were selected for micro-array analysis based on their origin and phenotypic and genotypic profiles. Strains with different antimicrobial resistance phenotypes and PFGE profiles from turkeys originating from the two main hatcheries were characterized using the Identibac S. aureus Genotyping DNA Microarray (Alere Technologies GmbH, Jena, Germany) according to the manufacturer's instructions. The DNA microarray covers 333 genetic markers for S. aureus, including species markers, capsule and agr group typing markers, as well as diverse antimicrobial resistance and virulence factors. A full list including primer and probe sequences is available online (http://identibac.com/en/home.html). Specific PCR assays were conducted for resistance and virulence genes not included in the microarray. Primer sequences and PCR conditions are shown in .
Table 1. Primers used in the present study.
Results
Characterization of S. aureus from diseased turkeys
The 34 isolates investigated in this work were distributed among nine spa types (t150, t034, t571, t588, t645, t899, t2164, t2970, t4652), with t034 being the most common type (). Most isolates (91.2%, 31/34) showed spa types related to the CC398 (t034, t571, t588, t899, t2970, t4652), and were confirmed as CC398 using the sau1-hsdS1 PCR assay. The three isolates with spa types not related to CC398 (t150, t645, t2164) were negative for the CC398 PCR and belonged to different CCs (). The control PFGE strain NCTC 8325 has spa type t211 and belongs to ST8 (CC8). Genomic DNA of the non-CC398 isolates and the control strain NCTC 8325 was digested with both SmaI and Cfr9I. In contrast, DNA of the CC398 isolates was resistant to SmaI and susceptible to Cfr9I restriction, yielding seven different profiles (). On the basis of the Dice similarity coefficient, a dendrogram with the Cfr9I-PFGE patterns was constructed (). At a similarity index of 71.7%, all PFGE profiles of CC398 isolates were grouped into a single cluster, while the non-CC398 isolates and the control NCTC 8325 PFGE strain were clearly separated in the dendrogram.
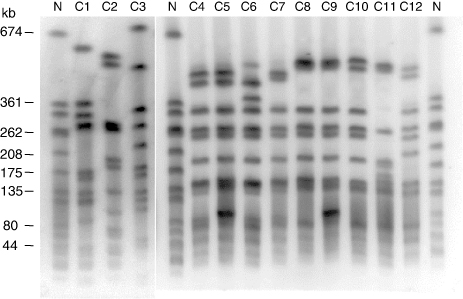
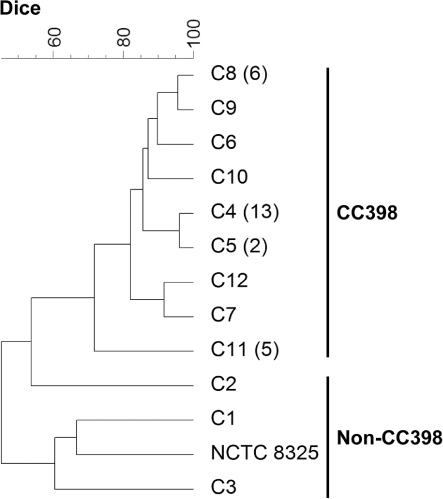
Table 2. Features of the turkey S. aureus isolates analysed in this study.
The isolates showed 11 resistance profiles. All strains were susceptible to kanamycin, gentamicin, linezolid, chloramphenicol, trimethoprim, vancomycin and mupirocin (). Most resistance profiles were represented by a single isolate, and only three profiles were shared by two or more CC398 isolates: R2 (16 isolates), R3 (seven isolates) and R5 (two isolates). The highest incidence of individual resistance among CC398 isolates corresponded to penicillin and tetracycline (100% each), followed by ciprofloxacin (32.4%), fusidic acid (16.1%), clindamycin (12.9%), erythromycin (9.7%), streptomycin and tiamulin (6.5% each), and quinupristin/dalfopristin (3.2%). Only one cefoxitin-resistant isolate (isolate 63-D10) with a high resistance to this antimicrobial (16 µg/µl) was found. The isolate was typed as MRSA by the triplex PCR and carried SCCmec IVa. The combination of the different typing techniques allowed the classification of the 34 isolates investigated in the present work into 21 different strains ().
Microarray typing and detection of resistance and virulence genes
Selected isolates (corresponding to strains 1, 2, 6, 8, 10, 11, 12, 15, 18, 19, 21 and 22) with different antimicrobial resistance phenotypes and PFGE profiles were characterized by microarray typing (). Most genes were homogeneously distributed in all strains, including typical S. aureus species marker and regulatory genes (23S-rRNA, gapA, katA, coA, spa, sbi, nuc, fnbA, vraS, eno, saeS), the accessory gene regulator agrI, capsule 5-related genes (capH5, capJ5, capK5), the biofilm production genes of the icaACD operon, genes encoding leukocidins (lukS-F, hlgA, lukX, lukY-variant 1), haemolysins (hla, hld), proteases (aur, sspA, sspB, sspP), adhesion factors (cflA, cflB, cna, ebh, eno, fib, ebpS, fnbA, fnbB, map, sdrC, vwb), immune-evasion factors (isaB, isdA, hysA1, hysA2), a putative transport protein (lmrP), a site-specific deoxyribonuclease subunit X (hsdSx), and staphylococcal superantigen-like proteins from the vSaα genomic islands (setB1, setB2, setB3, setC, ssl1 [set6], ssl2 [set7], ssl4 [set9/ssl4], ssl5 [set3/ssl5], ssl7 [set1/ssl7], ssl9 [set5/ssl9] and ssl10 [set4/ssl10]).
Table 3. Presence of antimicrobial resistance genes, virulence, adhesion and inmunoevasion factors by DNA microarray and PCR test in selected isolates.
All strains were penicillin and tetracycline resistant and carried the penicillin–ampicillin resistance operon bla (blaZ, blaI, and blaR) and the tetracycline resistance gene tetM; but three strains also carried the tetracycline resistance gene tetK. The erythromycin resistant strains 2, 18 and 19 carried the gene ermC, and the unique MRSA isolate (strain 22) carried the streptogramine resistance gene vgaA, the qacC efflux pump and some SCCmec genes (mecA, the truncated mecR, ugpQ, ccrA2 and ccrB2) related to SCCmec IV. All methicillin-susceptible S. aureus (MSSA) isolates possessed an intact beta-haemolysin gene (hlb), while the MRSA isolate (strain 22) harboured the hlb gene truncated after the probable insertion of the immune-evasion phage-borne genes sak (staphylokinase), chp (chemotaxis inhibitory protein) and scn (staphylococcal complement inhibitor). Moreover, the MRSA strain also carried the enterotoxin G gene (entG, also called seg), and other adhesion (bbp, sdrD) and immune evasion proteins (mprF), as well as the site-specific deoxyribonuclease subunit 2 (hsdS2).
The 34 isolates investigated in this work were also subjected to specific PCR assays for resistance and virulence genes not included in the microarray, with strain 29-G3 (strain 1) possessing the copper resistance gene copB, the isolate 30-D8 (strain 3) positive for the apramycin resistance gene ampA, and all isolates, except the MRSA strain 22, positive for the presence of φAvβ prophage avian-niche-specific genes SAAV_2008 and SAAV_2009. The remaining investigated genes were not detected.
Discussion
S. aureus is an opportunistic pathogen in food animals, and has been isolated from various farm animal species including pigs, poultry and cattle (Vanderhaeghen et al., Citation2012; Fluit, Citation2012). Most research performed among livestock isolates is focused on MRSA isolates, particularly from the lineage CC398, which is highly prevalent among farm animals. As such, little research on S. aureus from poultry has included MSSA isolates, despite their potential pathogenicity. In the present study, S. aureus strains, including both MRSA and MSSA, isolated from diseased turkeys from a region in France over a period of 5 years have been analysed. The results contribute to a better understanding of the epidemiology of S. aureus in this farm bird species.
The molecular characterization of the S. aureus isolates from diseased turkeys revealed that the vast majority belonged to CC398. The spa types detected in the isolates from this study were diverse, being types t034 and t899 previously reported among MRSA isolates from turkey products and healthy and diseased turkeys (Argudín et al., Citation2010a; Bystroń et al., Citation2010; Feßler et al., Citation2011a; Richter et al., Citation2012; Monecke et al., Citation2013). The spa type t034 is one of the most common spa types found in CC398 isolates of diverse origin, while t899 has been found in pig isolates (van Duijkeren et al., Citation2008), as a cause of infection in pig farmers (Pan et al., Citation2009) and recently in healthy carrier chickens in Belgium (Nemeghaire et al., Citation2013). Interestingly, the spa type t899 has also been reported in MRSA from CC9 (Wagenaar et al., Citation2009). In fact, comparative genomic analysis with other STs suggested that the region surrounding the spa gene in t899 isolates was acquired horizontally from an ST9 donor (Price et al., Citation2012). Direct phylogenetic analysis demonstrated t899 as the first lineage to diverge from the other CC398 isolates, but with the exclusion of the single nucleotide polymorphisms in the region surrounding the spa gene, the t899 lineage clustered with a more divergent clade of European isolates and a clade of human-associated MSSA isolates from France and French Guiana (Price et al., Citation2012). The remaining spa types found in this study have been previously reported for isolates from pigs (t571 and t2970; Argudín et al., Citation2010a), pig farmers and horses (t588; van Duijkeren et al., Citation2010; Lozano et al., Citation2011) or broiler flocks (t4652; Mulders et al., Citation2010). The three non-CC398 isolates had spa types that have been found before in pathogenic human MSSA isolates in Europe (Deurenberg et al., Citation2009; Pérez-Vázquez et al., Citation2009; Rijnders et al., Citation2012) and they were associated with the human lineages CC5, CC101 and CC121.
The different antimicrobial usages between countries and animal species can influence the antimicrobial resistance patterns observed in bacterial populations. The S. aureus isolates analysed in this study showed rather homogeneous resistance profiles, and about one-half showed a multi-resistance pattern (resistance to three or more antimicrobials). Most strains were MSSA, and showed typical resistance traits (penicillin and tetracycline resistance) of both MRSA and MSSA CC398 isolates from different origins (Pantosti, Citation2012, Crombé et al., Citation2013). Interestingly, the isolates from this study showed low resistance percentages to erythromycin (8.8%) and clindamycin (11.8%), while other studies, particularly those on S. aureus from turkey flocks, turkey meat and products from Germany, have recorded high percentages of erythromycin and/or clindamycin resistance, up to 87%. However, these strains were MRSA and had the typical MRSA CC398 multi-resistance pattern (Feßler et al., Citation2011a; Richter et al., Citation2012; Monecke et al., Citation2013). The MRSA CC398 strain isolated in this study showed a similar multi-resistance pattern, but also amongst MSSA from turkeys, high percentages of erythromycin resistance (93%) were found in Germany (Monecke et al., Citation2013). Similarly, in MRSA isolates from broiler chickens in Belgium (Nemeghaire et al., Citation2013) and the Netherlands (Wendlandt et al., Citation2013) high resistance percentages (up to 50%) to macrolides and lincosamides were reported. Conversely, a study in earlier poultry S. aureus Belgian isolates (recovered between 1970 and 1972) showed a lower percentage of resistance (<20%) to these antimicrobials (Nemati et al., Citation2008). Regarding the other antimicrobials examined, the prevalence of resistance to ciprofloxacin (32.4%) was similar to a study of MRSA isolates from birds and staff in flocks of turkeys examined in Germany (Richter et al., Citation2012). Other studies in Germany showed fluoroquinolone resistance percentages of between 13 and 59% in MRSA isolates (Feßler et al., Citation2011a; Monecke et al., Citation2013) and of 86% in MSSA isolates (Monecke et al., Citation2013). Lower fluoroquinolone resistance percentages (about 17%) were also recorded among broiler chickens in the Netherlands (Wendlandt et al., Citation2013) and MRSA CC398 from different origin in Belgium (Jamrozy et al., Citation2012). These country and regional variations underlie the importance of the antimicrobial resistance monitoring studies to determine and substantiate treatment regimens and strategies.
Since its discovery, the CC398 lineage has been considered a livestock-associated commensal, but recent findings based on whole genome sequencing (Price et al., Citation2012) suggest that this lineage originated from humans and spread to livestock. Microarray and sequence-based studies support that CC398 isolates essentially carry the same core genome and have different secreted genes located on mobile genetic elements depending on the host and country of origin (Stegger et al., Citation2010; McCarthy et al., Citation2011, Citation2012; Price et al., Citation2012). For example, the φSa3 prophage genes (sak, chp and scn) were detected only in the typical human isolates and not in those from pigs or humans in contact with pigs (McCarthy et al., Citation2011, Citation2012). In the study by Price et al. (Citation2012), different livestock isolates from 19 countries were included and the loss of these human-niche-specific genes has also been confirmed for CC398 turkey isolates. Moreover, these turkey isolates have acquired the avian-niche-specific genes from the φAvβ prophage, which also belongs to the accessory gene pool of broiler chicken-associated S. aureus ST5 (Price et al., Citation2012). In accordance with this observation, this study demonstrated the presence of the avian-niche-specific genes SAAV_2008 and SAAV_2009 from the φAvβ prophage in all MSSA isolates including both CC398 and non-CC398 isolates. However, the φAvβ prophage is an hlb-converting phage (Lowder et al., Citation2009) and the MSSA strains investigated in this study carried an intact hlb gene. This indicates the possibility of a new non-hlb-converting phage carrying the specific SAAV_2008 and SAAV_2009 genes or another location for these determinants. This possibility has already been described for sak-containing prophages. The sak gene is usually related to φSa3 hlb-converting phages, which also carry chp and scn genes (Goerke et al., Citation2009); but a non-hlb-converting phage (φ6390) with sak has also been described (Goerke et al., Citation2006). Similar to the sak gene, the SAAV_2008 and SAAV_2009 determinants could also be located in a new non-hlb converting phage. The MRSA strain 22 was negative for the presence of the φAvβ prophage genes, but contained the three typical φSa3 prophage genes sak, chp and scn. These results differed from the study of Monecke et al. (Citation2013), where none of the CC398 S. aureus strains isolates from diseased turkeys in Germany carried the φSa3 prophage genes chp and scn, but 65% of the MSSA and 9% of the MRSA strains examined carried the staphylokinase gene sak together with the enterotoxin gene sea, which is also associated with φSa3 prophages (Monecke et al., Citation2013). Interestingly, the microarray results on German turkey isolates provided by Monecke et al. (Citation2013) included isolates with intact hlb together with scn or sak and scn, indicating the possibility of other new non hlb-converting phages. The carriage of other virulence factors (including leukocidins, proteases, staphylococcal superantigen-like proteins and the haemolysin genes hla and hld), adhesion and immune-evasion factors were similar between the strains of this study and those of other German turkey isolates (Monecke et al., Citation2013). Interestingly, the core genome seems stable, and these turkey isolates have a similar genetic background to CC398 isolates from poultry, pigs, bovines and horses (Nemati et al., Citation2009; Walther et al., Citation2009; Hallin et al., Citation2011; Jamrozy et al., Citation2012). Only minor differences were detected, particularly among the CC398 isolates from this study that were negative for the presence of bbp and sdrD genes, while the MRSA strain 22 carried both genes. These genes might be important for S. aureus pathogenicity in a human host. The Bbp protein is important in the invasion of bone tissue and thus might be of relevance in the pathogenicity of osteomyelitis (Tung et al., Citation2000), while SdrD might play a role in the interactions between the pathogen and the human immune system and serum or may specifically react to nutrients and other factors present in human blood (Sitkiewicz et al., Citation2011). The absence of both genes (bbp and sdrD) was also common in German MSSA turkey isolates (Monecke et al., Citation2013), but also in Belgian MRSA isolates from poultry, pigs, horses, chicken and bovines (Nemati et al., Citation2009; Jamrozy et al., Citation2012). Conversely, these genes appeared common among German MRSA isolates from poultry (Monecke et al., Citation2013) and swine (Kadlec et al., Citation2009). These results confirm that the mobile genetic elements distributed among CC398 isolates are influenced by host and country.
The unique MRSA isolate (strain 22) carried the enterotoxin G gene, which is carried by the egc cluster of the vSaß genomic island (Argudín et al., Citation2010b). The egc cluster is considered a nursery of enterotoxins and enterotoxin-like genes, and some variants developed through recombination processes have been described (Argudín et al., Citation2010b). Interestingly, the enterotoxin genes of the egc locus are usually more common among nasal than invasive isolates, and it has been proposed that the egc-encoded toxins could enhance the potential carriage of S. aureus (van Belkum et al., Citation2006). An incomplete egc cluster, with only the seg gene, has also been described in one human nasal isolate in Japan (Omoe et al., Citation2005) and one ST398 MRSA from pig in France (Laurent et al., Citation2009).
This study provides evidence that S. aureus CC398 has an extraordinary ability to adapt from one reservoir to another, and highlights the need for further surveillance and research on this potentially versatile and pathogenic pathogen.
Acknowledgements
This research was funded by the CODA-CERVA. M. A. Argudín is supported by a research grant from the Fundación Alfonso Martín Escudero.
References
- Alfonso, M. & Barnes, H.J. (2006). Neonatal osteomyelitis associated with Staphylococcus aureus in turkey poults. Avian Diseases, 50, 148–151. 10.1637/7298-110104R.1
- Andreasen, C.B. (2003). Staphylococcosis. In Y.M. Saif, H.J. Barnes, J.R. Glisson, A.M. Fadly, L.R. McDougald & D.E. Swayne (Eds.) Diseases of Poultry 11th edn (pp. 797–804). Ames: Iowa State University Press.
- Argudín, M.A., Fetsch, A., Tenhagen, B.A., Hammerl, J.A., Hertwig, S., Kowall, J., Rodicio, M.R., Käsbohrer, A., Helmuth, R., Schroeter, A., Mendoza, M.C., Bräunig, J., Appel, B. & Guerra, B. (2010a). High heterogeneity within methicillin-resistant Staphylococcus aureus ST398 isolates, defined by Cfr9I macrorestriction-pulsed-field gel electrophoresis profiles and spa and SCCmec types. Applied and Environmental Microbiology, 76, 652–658. 10.1128/AEM.01721-09
- Argudín, M.A., Mendoza, M.C. & Rodicio, M.R. (2010b). Food poisoning and Staphylococcus aureus enterotoxins. Toxins, 2, 1751–1773. 10.3390/toxins2071751
- Argudín, M.A., Rodicio, M.R. & Guerra, B. (2010c). The emerging methicillin-resistant Staphylococcus aureus ST398 clone can easily be typed using the Cfr9I SmaI-neoschizomer. Letters in Applied Microbiology, 50, 127–130. 10.1111/j.1472-765X.2009.02742.x
- Argudín, M.A., Tenhagen, B.A., Fetsch, A., Sachsenröder, J., Käsbohrer, A., Schroeter, A., Hammerl, J.A., Hertwig, S., Helmuth, R., Bräunig, J., Mendoza, M.C., Appel, B., Rodicio, M.R. & Guerra, B. (2011). Virulence and resistance determinants of German Staphylococcus aureus ST398 isolates from nonhuman sources. Applied and Environmental Microbiology, 77, 3052–3060. 10.1128/AEM.02260-10
- Bystroń, J., Podkowik, M., Piasecki, T., Wieliczko, A., Molenda, J. & Bania, J. (2010). Genotypes and enterotoxin gene content of S. aureus isolates from poultry. Veterinary Microbiology, 144, 498–501. 10.1016/j.vetmic.2010.01.029
- Cavaco, L.M., Hasman, H., Stegger, M., Andersen, P.S., Skov, R., Fluit, A.C., Ito, T. & Aarestrup, F.M. (2010). Cloning and occurrence of czrC, a gene conferring cadmium and zinc resistance in methicillin-resistant Staphylococcus aureus CC398 isolates. Antimicrobial Agents and Chemotherapy, 54, 3605–3608. 10.1128/AAC.00058-10
- Corrand, L., Lucas, M.N., Douet, J.Y., Etienne, C.L., Albaric, A.O., Cadec, A. & Guérin, J.L. (2012). A case of unilateral periorbital cellulitis and mandibular osteomyelitis in a turkey flock. Avian Diseases, 56, 427–431. 10.1637/9910-083111-Case.1
- Crombé, F., Argudín, M.A., Vanderhaeghen, W., Hermans, K., Haesebrouck, F. & Butaye, P. (2013). Transmission dynamics of methicillin-resistant Staphylococcus aureus in pigs. Frontiers in Microbiology, 4, 57.
- de Boer, E., Zwartkruis-Nahuis, J.T., Wit, B., Huijsdens, X.W., de Neeling, A.J., Bosch, T., van Oosterom, R.A., Vila, A. & Heuvelink, A.E. (2009). Prevalence of methicillin-resistant Staphylococcus aureus in meat. International Journal of Food Microbiology, 134, 52–56. 10.1016/j.ijfoodmicro.2008.12.007
- Deurenberg, R.H., Rijnders, M.I., Sebastian, S., Welling, M.A., Beisser, P.S. & Stobberingh, E.E. (2009). The Staphylococcus aureus lineage-specific markers collagen adhesin and toxic shock syndrome toxin 1 distinguish multilocus sequence typing clonal complexes within spa clonal complexes. Diagnostic Microbiology and Infectious Disease, 65, 116–122. 10.1016/j.diagmicrobio.2009.07.007
- Devriese, L.A. (1980). Pathogenic staphylococci in poultry. Journal of Applied Bacteriology, 50, 357.
- Feßler, A.T., Kadlec, K., Hassel, M., Hauschild, T., Eidam, C., Ehricht, R., Monecke, S. & Schwarz, S. (2011a). Characterization of methicillin-resistant Staphylococcus aureus isolates from food and food products of poultry origin in Germany. Applied and Environmental Microbiology, 77, 7151–7157. 10.1128/AEM.00561-11
- Feßler, A.T., Kadlec, K. & Schwarz, S. (2011b). Novel apramycin resistance gene apmA in bovine and porcine methicillin-resistant Staphylococcus aureus ST398 isolates. Antimicrobial Agents and Chemotherapy, 55, 373–375. 10.1128/AAC.01124-10
- Feßler, A., Scott, C., Kadlec, K., Ehricht, R., Monecke, S. & Schwarz, S. (2010). Characterization of methicillin-resistant Staphylococcus aureus ST398 from cases of bovine mastitis. Journal of Antimicrobial Chemotherapy, 65, 619–625. 10.1093/jac/dkq021
- Fitzgerald, J.R. (2012). Livestock-associated Staphylococcus aureus: origin, evolution and public health threat. Trends in Microbiology, 20, 192–198. 10.1016/j.tim.2012.01.006
- Fluit, A.C. (2012). Livestock-associated Staphylococcus aureus. Clinical Microbiology and Infection, 18, 735–744. 10.1111/j.1469-0691.2012.03846.x
- Goerke, C., Pantucek, R., Holtfreter, S., Schulte, B., Zink, M., Grumann, D., Bröker, B.M., Doskar, J. & Wolz, C. (2009). Diversity of prophages in dominant Staphylococcus aureus clonal lineages. Journal of Bacteriology, 191, 3462–3468. 10.1128/JB.01804-08
- Goerke, C., Wirtz, C., Flückiger, U. & Wolz, C. (2006). Extensive phage dynamics in Staphylococcus aureus contributes to adaptation to the human host during infection. Molecular Microbiology, 61, 1673–1685. 10.1111/j.1365-2958.2006.05354.x
- Hallin, M., De Mendonça, R., Denis, O., Lefort, A., El Garch, F., Butaye, P., Hermans, K. & Struelens, M.J. (2011). Diversity of accessory genome of human and livestock-associated ST398 methicillin resistant Staphylococcus aureus strains. Infection, Genetics and Evolution, 11, 290–299. 10.1016/j.meegid.2010.10.021
- Jamrozy, D.M., Fielder, M.D., Butaye, P. & Coldham, N.G. (2012). Comparative genotypic and phenotypic characterisation of methicillin-resistant Staphylococcus aureus ST398 isolated from animals and humans. PLoS ONE, 7, e40458. 10.1371/journal.pone.0040458
- Kadlec, K., Ehricht, R., Monecke, S., Steinacker, U., Kaspar, H., Mankertz, J. & Schwarz, S. (2009). Diversity of antimicrobial resistance pheno- and genotypes of methicillin-resistant Staphylococcus aureus ST398 from diseased swine. Journal of Antimicrobial Chemotherapy, 64, 1156–1164. 10.1093/jac/dkp350
- Köck, R., Schaumburg, F., Mellmann, A., Köksal, M., Jurke, A., Becker, K. & Friedrich, A.W. (2013). Livestock-associated methicillin-resistant Staphylococcus aureus (MRSA) as causes of human infection and colonization in Germany. PLoS ONE, 8, e55040. 10.1371/journal.pone.0055040
- Kondo, Y., Ito, T., Ma, X.X., Watanabe, S., Kreiswirth, B.N., Etienne, J. & Hiramatsu, K. (2007). Combination of multiplex PCRs for staphylococcal cassette chromosome mec type assignment: rapid identification system for mec, ccr, and major differences in junkyard regions. Antimicrobial Agents and Chemotherapy, 51, 264–274. 10.1128/AAC.00165-06
- Laurent, F., Jouy, E., Granier, S., BES, M., Ruimy, R., Felix, B., Le Roux, A., Sanders, P., Chauvin, C., Etienne, J. & Brisabois, A. (2009). Molecular characterization of antimicrobial resistance genes and virulence genes by using microarrays in representative ST398 MRSA isolates from pigs in France. Proceedings of the ASM-ESCMID Conference on Methicillin-resistant Staphylococci in Animals: Veterinary and Public Health Implications (p. 25). London, UK.
- Lindsay, J.A. (2010). Genomic variation and evolution of Staphylococcus aureus. International Journal of Medical Microbiology, 300, 98–103. 10.1016/j.ijmm.2009.08.013
- Lowder, B.V., Guinane, C.M., Ben Zakour, N.L., Weinert, L.A., Conway-Morris, A., Cartwright, R.A., Simpson, A.J., Rambaut, A., Nübel, U. & Fitzgerald, J.R. (2009). Recent human-to-poultry host jump, adaptation, and pandemic spread of Staphylococcus aureus. Proceedings of the National Academy of Sciences USA, 106, 19545–19550. 10.1073/pnas.0909285106
- Lozano, C., Aspiroz, C., Ara, M., Gómez-Sanz, E., Zarazaga, M. & Torres, C. (2011). Methicillin-resistant Staphylococcus aureus (MRSA) ST398 in a farmer with skin lesions and in pigs of his farm: clonal relationship and detection of lnu(A) gene. Clinical Microbiology and Infection, 17, 923–927. 10.1111/j.1469-0691.2010.03437.x
- Maes, N., Magdalena, J., Rottiers, S., De Gheldre, Y. & Struelens, M.J. (2002). Evaluation of a triplex PCR assay to discriminate Staphylococcus aureus from coagulase-negative staphylococci and determine methicillin resistance from blood cultures. Journal of Clinical Microbiology, 40, 1514–1517. 10.1128/JCM.40.4.1514-1517.2002
- Malbruny, B., Werno, A.M., Murdoch, D.R., Leclercq, R. & Cattoir, V. (2011). Cross-resistance to lincosamides, streptogramins A, and pleuromutilins due to the lsa(C) gene in Streptococcus agalactiae UCN70. Antimicrobial Agents and Chemotherapy, 55, 1470–1474. 10.1128/AAC.01068-10
- McCarthy, A.J., van Wamel, W., Vandendriessche, S., Larsen, J., Denis, O., Garcia-Graells, C., Uhlemann, A.C., Lowy, F.D., Skov, R. & Lindsay, J.A. (2012). Staphylococcus aureus CC398 clade associated with human-to-human transmission. Applied and Environmental Microbiology, 78, 8845–8848. 10.1128/AEM.02398-12
- McCarthy, A.J., Witney, A.A., Gould, K.A., Moodley, A., Guardabassi, L., Voss, A., Denis, O., Broens, E.M., Hinds, J. & Lindsay, J.A. (2011). The distribution of mobile genetic elements (MGEs) in MRSA CC398 is associated with both host and country. Genome Biology and Evolution, 3, 1164–1174. 10.1093/gbe/evr092
- Monecke, S., Ruppelt, A., Wendlandt, S., Schwarz, S., Slickers, P., Ehricht, R. & Jäckel, S.C. (2013). Genotyping of Staphylococcus aureus isolates from diseased poultry. Veterinary Microbiology, 162, 806–812. 10.1016/j.vetmic.2012.10.018
- Mulders, M.N., Haenen, A.P., Geenen, P.L., Vesseur, P.C., Poldervaart, E.S., Bosch, T., Huijsdens, X.W., Hengeveld, P.D., Dam-Deisz, W.D., Graat, E.A., Mevius, D., Voss, A. & Van De Giessen, A.W. (2010). Prevalence of livestock-associated MRSA in broiler flocks and risk factors for slaughterhouse personnel in The Netherlands. Epidemiology and Infection, 138, 743–755. 10.1017/S0950268810000075
- Nemati, M., Hermans, K., Devriese, L.A., Maes, D. & Haesebrouck, F. (2009) Screening of genes encoding adhesion factors and biofilm formation in Staphylococcus aureus isolates from poultry. Avian Pathology, 38, 513–517. 10.1080/03079450903349212
- Nemati, M., Hermans, K., Lipinska, U., Denis, O., Deplano, A., Struelens, M., Devriese, L.A., Pasmans, F. & Haesebrouck, F. (2008). Antimicrobial resistance of old and recent Staphylococcus aureus isolates from poultry: first detection of livestock-associated methicillin-resistant strain ST398. Antimicrobial Agents and Chemotherapy, 52, 3817–3819. 10.1128/AAC.00613-08
- Nemeghaire, S., Roelandt, S., Argudín, M.A., Haesebrouck, F. & Butaye, P. (2013). Characterization of methicillin-resistant Staphylococcus aureus from healthy carrier chickens. Avian Pathology, 42, 342–346. 10.1080/03079457.2013.805183
- Ng, L.K., Martin, I., Alfa, M. & Mulvey, M. (2001). Multiplex PCR for the detection of tetracycline resistant genes. Molecular and Cellular Probes, 15, 209–215. 10.1006/mcpr.2001.0363
- Omoe, K., Hu, D.L., Takahashi-Omoe, H., Nakane, A. & Shinagawa, K. (2005). Comprehensive analysis of classical and newly described staphylococcal superantigenic toxin genes in Staphylococcus aureus isolates. FEMS Microbiology Letters, 246, 191–198. 10.1016/j.femsle.2005.04.007
- Pan, A., Battisti, A., Zoncada, A., Bernieri, F., Boldini, M., Franco, A., Giorgi, M., Iurescia, M., Lorenzotti, S., Martinotti, M., Monaci, M. & Pantosti, A. (2009). Community-acquired methicillin-resistant Staphylococcus aureus ST398 infection, Italy. Emerging Infectious Diseases, 15, 845–847. 10.3201/eid1505.081417
- Pantosti, A. (2012). Methicillin-Resistant Staphylococcus aureus associated with animals and its relevance to human health. Frontiers in Microbiology, 3, 127. 10.3389/fmicb.2012.00127
- Pérez-Vázquez, M., Vindel, A., Marcos, C., Oteo, J., Cuevas, O., Trincado, P., Bautista, V., Grundmann, H., Campos, J. & EARSS Spain spa-typing Group. (2009). Spread of invasive Spanish Staphylococcus aureus spa-type t067 associated with a high prevalence of the aminoglycoside-modifying enzyme gene ant(4')-Ia and the efflux pump genes msrA/msrB. Journal of Antimicrobial Chemotherapy, 63, 21–31. 10.1093/jac/dkp078
- Price, L.B., Stegger, M., Hasman, H., Aziz, M., Larsen, J., Andersen, P.S., Pearson, T., Waters, A.E., Foster, J.T., Schupp, J., Gillece, J., Driebe, E., Liu, C.M., Springer, B., Zdovc, I., Battisti, A., Franco, A., Zmudzki, J., Schwarz, S., Butaye, P., Jouy, E., Pomba, C., Porrero, M.C., Ruimy, R., Smith, T.C., Robinson, D.A., Weese, J.S., Arriola, C.S., Yu, F., Laurent, F., Keim, P., Skov, R. & Aarestrup, F.M. (2012). Staphylococcus aureus CC398: host adaptation and emergence of methicillin resistance in livestock. MBio, 21, 3.
- Richter, A., Sting, R., Popp, C., Rau, J., Tenhagen, B.A., Guerra, B., Hafez, H.M. & Fetsch, A. (2012). Prevalence of types of methicillin-resistant Staphylococcus aureus in turkey flocks and personnel attending the animals. Epidemiology and Infection, 10, 1–10.
- Rijnders, M.I., Wolffs, P.F., Hopstaken, R.M., den Heyer, M., Bruggeman, C.A. & Stobberingh, E.E. (2012). Spread of the epidemic European fusidic acid-resistant impetigo clone (EEFIC) in general practice patients in the south of The Netherlands. Journal of Antimicrobial Chemotherapy, 67, 1176–1180. 10.1093/jac/dkr590
- Schwendener, S. & Perreten, V. (2011). New transposon Tn6133 in methicillin-resistant Staphylococcus aureus ST398 contains vga(E), a novel streptogramin A, pleuromutilin, and lincosamide resistance gene. Antimicrobial Agents and Chemotherapy, 55, 4900–4904. 10.1128/AAC.00528-11
- Sitkiewicz, I., Babiak, I. & Hryniewicz, W. (2011). Characterization of transcription within sdr region of Staphylococcus aureus. Antonie Van Leeuwenhoek, 99, 409–416. 10.1007/s10482-010-9476-7
- Stegger, M., Lindsay, J.A., Moodley, A., Skov, R., Broens, E.M. & Guardabassi, L. (2011). Rapid PCR detection of Staphylococcus aureus clonal complex 398 by targeting the restriction-modification system carrying sau1-hsdS1. Journal of clinical Microbiology, 49, 732–734. 10.1128/JCM.01970-10
- Stegger, M., Lindsay, J.A., Sørum, M., Gould, K.A. & Skov, R. (2010). Genetic diversity in CC398 methicillin-resistant Staphylococcus aureus isolates of different geographical origin. Clinical Microbiology and Infection, 16, 1017–1019.
- Tung, Hs., Guss, B., Hellman, U., Persson, L., Rubin, K. & Rydén, C. (2000). A bone sialoprotein-binding protein from Staphylococcus aureus: a member of the staphylococcal Sdr family. Biochemical Journal, 345, 611–619. 10.1042/0264-6021:3450611
- van Belkum, A., Melles, D.C., Snijders, S.V., van Leeuwen, W.B., Wertheim, H.F., Nouwen, J.L., Verbrugh, H.A. & Etienne, J. (2006). Clonal distribution and differential occurrence of the enterotoxin gene cluster, egc, in carriage- versus bacteremia-associated isolates of Staphylococcus aureus. Journal of Clinical Microbiology, 44, 1555–1557. 10.1128/JCM.44.4.1555-1557.2006
- Vanderhaeghen, W., Cerpentier, T., Adriaensen, C., Vicca, J., Hermans, K. & Butaye, P. (2010). Methicillin-resistant Staphylococcus aureus (MRSA) ST398 associated with clinical and subclinical mastitis in Belgian cows. Veterinary Microbiology, 144, 166–171. 10.1016/j.vetmic.2009.12.044
- Vanderhaeghen, W., Hermans, K., Haesebrouck, F. & Butaye P. (2012). Methicillin-resistant Staphylococcus aureus (MRSA) in food production animals. Epidemiology and Infection, 138, 606–625. 10.1017/S0950268809991567
- van Duijkeren, E., Ikawaty, R., Broekhuizen-Stins, M.J., Jansen, M.D., Spalburg, E.C., de Neeling, A.J., Allaart, J.G., van Nes, A., Wagenaar, J.A. & Fluit, A.C. (2008). Transmission of methicillin-resistant Staphylococcus aureus strains between different kinds of pig farms. Veterinary Microbiology, 126, 383–389. 10.1016/j.vetmic.2007.07.021
- van Duijkeren, E., Moleman, M., Sloet van Oldruitenborgh-Oosterbaan, M.M., Multem, J., Troelstra, A., Fluit, A.C., van Wamel, W.J., Houwers, D.J., de Neeling, A.J. & Wagenaar, J.A. (2010). Methicillin-resistant Staphylococcus aureus in horses and horse personnel: an investigation of several outbreaks. Veterinary Microbiology, 141, 96–102. 10.1016/j.vetmic.2009.08.009
- Wagenaar, J.A., Yue, H., Pritchard, J., Broekhuizen-Stins, M., Huijsdens, X., Mevius, D.J., Bosch, T. & van Duijkeren, E. (2009). Unexpected sequence types in livestock associated methicillin-resistant Staphylococcus aureus (MRSA): MRSA ST9 and a single locus variant of ST9 in pig farming in China. Veterinary Microbiololgy, 139, 405–409. 10.1016/j.vetmic.2009.06.014
- Walther, B., Monecke, S., Ruscher, C., Friedrich, A.W., Ehricht, R., Slickers, P., Soba, A., Wleklinski, C.G., Wieler, L.H. & Lübke-Becker, A. (2009). Comparative molecular analysis substantiates zoonotic potential of equine methicillin-resistant Staphylococcus aureus. Journal of Clinical Microbiology 47, 704–710. 10.1128/JCM.01626-08
- Wendlandt, S., Kadlec, K., Feßler, A.T., Monecke, S., Ehricht, R., van de Giessen, A.W., Hengeveld, P.D., Huijsdens, X., Schwarz, S. & van Duijkeren, E. (2013). Resistance phenotypes and genotypes of methicillin-resistant Staphylococcus aureus isolates from broiler chickens at slaughter and abattoir workers. Journal of Antimicrobial Chemotherapy, 68, 2458–2463.