Abstract
In the present study a well-characterized strain collection (n = 33) of Avibacterium species was investigated by matrix-assisted laser desorption ionization–time-of flight mass spectrometry (MALDI-TOF MS). The robustness of the currently available reference database (Bruker Biotyper 3.0) was tested to determine the degree of identification of these strains. Reproducible signal patterns were obtained from all strains. However, identification of most strains was only possible at genus level. Furthermore, two strains could not be identified by this method. Based on their protein spectra profiles, a MALDI main spectra dendrogram was created to determine their relationship. Most strains were closely related—for example, 26 strains formed cluster 1 including the type strains of Avibacterium volantium, Avibacterium gallinarum, Avibacterium endocarditidis and Avibacterium avium—while Avibacterium paragallinarum biovars 1 and 2 formed cluster 2 and, finally, strain 55000 remained on its own. The present MALDI-TOF MS results confirm recent findings that only certain isolates of Av. paragallinarum represent a well-defined species within the genus Avibacterium, making a taxonomic revision essential. To improve identification of Avibacterium at species level by MALDI-TOF MS, relevant reference strains were included in the newly created database and results are presented. In conclusion, Av. paragallinarum can be identified by MALDI/Biotyper and not the other species of the genus.
Introduction
Within the Pasteurellaceae family, the genus Avibacterium comprises five validly named species—Avibacterium gallinarum, Av. paragallinarum, Av. avium, Av. volantium and Av. endocarditidis—and one unnamed taxon, Avibacterium species A (Blackall et al., Citation2005; Bisgaard et al., Citation2007). Correct characterization and reliable identification of the members of this group, however, still represent a major challenge in diagnostic laboratories, with misidentification as a frequent and a serious problem. Some isolates of Avibacterium are difficult to classify based upon phenotypic identification due to variable species characteristics (Blackall, Citation1988; Blackall & Norskov-Lauritsen, Citation2008). Additionally, Bisgaard et al. (Citation2012) showed that genotyping, including DNA sequencing, did not result in unambiguous identification of these bacteria. The authors speculated that the close phylogenetic relationship and the high similarity between most isolates investigated, except for Av. paragallinarum, may be due to the fact that incipient species are present. Another drawback for unambiguous classification is the fact that some species descriptions are based on only a few isolates just as polyphyly was shown in some species (Mutters et al., Citation1985; Blackall et al., Citation2005).
The present investigation based on matrix-assisted laser desorption ionization–time-of flight mass spectrometry (MALDI-TOF MS) reports a new approach to analyse all known Avibacterium species by means of mass spectral fingerprinting. Detection of protein profiles has become a convenient tool for the rapid analysis of bacteria and has already been implemented in routine diagnostic laboratories (Freiwald & Sauer, Citation2009; Seng et al., Citation2009; Sauer & Kliem, Citation2010). MALDI-TOF MS identification of bacteria rests upon a library-based strategy in which spectra of unknown bacteria are compared with libraries containing spectra of known, reference bacteria. The Biotyper 3.0 reference database from Bruker Daltonics (Bremen, Germany) used in the present investigation contains four representatives of the genus Avibacterium: Av. endocarditidis DSM 18224T, Av. avium DSM 18557T, Av. gallinarum DSM 17481T and Av. volantium DSM 18578T. Kuhnert et al. (Citation2012) demonstrated that Av. paragallinarum could be clearly identified and separated from other taxa by MALDI-TOF MS, but in that study only one species of Avibacterium genus was investigated. In the present study, a well-characterized strain collection of all known species belonging to the Avibacterium genus was analysed by MALDI-TOF MS to determine the identification level of these strains based on the currently available spectra, and to investigate the relationship between the strains. Furthermore, the relationship of the strains was investigated by generating a score-oriented main spectra (MSP) dendrogram.
Materials and Methods
Bacterial isolates
The 33 Avibacterium strains analysed are presented in , representing 24 field isolates and nine type or reference strains. Phenotypic and genotypic results associated with these strains can be obtained from Bisgaard et al. (Citation2012). All strains were grown on PolyViteX agar (BioMeriéux, Vienna, Austria). The strains were incubated at 37°C for 48 h under microaerobic conditions (Genbox microaer; BioMeriéux).
Table 1. Avibacterium strains analysed: sequence type, identification results by phenotype and the Bruker MALDI/Biotyper database version 3 updated with our six reference strains.
Matrix-assisted laser desorption ionization–time-of flight mass spectrometry
Sample preparation for MALDI-TOF MS was performed as previously described in detail (Alispahic et al., Citation2010). Bacterial acid-soluble proteins were extracted using formic acid (70%) and acetonitrile according to the standard protocol from Bruker (Daltonics GmbH, Bremen, Germany). One micolitre of each bacterial extract was spotted eight times onto the MALDI target plate and air dried. Afterwards, 2 µl matrix solution (alpha-cyano-4-hydroxycinnamic acid in 50% acetonitrile/2.5% trifluoroacetic acid) were put on top of each sample and dried again. The parameter settings for the Microflex LT instrument were as follows: IS1, 20.08 kV; IS2, 16.77 kV; lens, 7.03 kV; detector gain, 1634 V. Two hundred and forty laser shots in 40 shot steps (in the linear, positive ion mode with a 60 Hz nitrogen laser from different positions of the target spot) were summarized automatically with the AutoXecute acquisition control software (Flex control 3.3; Bruker Daltonics). For database construction, each spot was measured three times ending with 24 spectra for each strain. All steps were performed at room temperature.
FlexAnalysis (version 3.3) software (Bruker Bruker Daltonics GmbH, Bremen, Germany) was used for visual inspection of the mass spectra. For automated data analysis, the raw spectra for unknown bacteria were processed using MALDI Biotyper software (Bruker Daltonics GmbH, Bremen, Germany) with the default settings. The software performs smoothing, normalization, baseline subtraction, and peak picking, thereby creating a list of the most significant peaks (m/z values) of the spectrum.
The MALDI Biotyper output is a log(score) in the range 0 to 3.0, computed by comparison of the peak list for an unknown isolate with the reference MSP in the database. A log(score) value between 1.7 and 2.0 represents genus identification, while a score value ≥ 2.0 represents identification at species level. Anything less than 1.7 was rated as non-identifiable by the software.
For creating a new database entry, mass spectra were processed with the software functionality and standard settings. The spectral peak lists for a particular strain were transferred into MSP containing information on average peak masses, average peak intensities and peak frequencies.
For strain relationship visualization, a dendrogram was formed based on MSP. Similar MSP result in a high matching score value. Each of these MSP is compared with all MSP of the analysed set. The list of score values is used to calculate normalized distance values between the analysed species, resulting in a matrix of matching scores. The visualization of the respective relationship between the MSP is displayed in a dendrogram using the following settings of the MALDI Biotyper 3.0 software: distance measure was set at correlation, linkage at average and score threshold value for a single organism at 700. Based on Sauer et al. (Citation2008), clusters of strains with distance levels <500 were classified as species.
Results
A reproducible signal pattern was obtained from all 33 Avibacterium strains investigated by MALDI-TOF MS. Signal patterns obtained were compared with data in the Bruker Biotyper reference database version 3 before it was updated with the six reference strains (data not shown). Reliable identification at genus level (score value 1.7 to 2.0) was demonstrated for 25 strains (76%). Identification at species level was only possible for six strains (18%): CCUG 18732, CCUG 18407, PG178, CCUG 12391T, CCUG 12833T and CCUG 3713T, all of which were identified as Av. endocarditidis. Two strains (CCUG 18396 and 55000) remained undiagnosed due to low scores value. Three strains of Av. endocarditidis Sequence Type 1, including the type strain, were only identified at genus level or remained unidentified. The type strains of Av. gallinarum, Av. volantium and Av. avium were identified as Av. endocarditidis (data not shown).
To determine the relationship between the 33 Avibacterium strains, a score-oriented MSP dendrogram was generated (). Based on a distance level of 500, three groups were present. The largest cluster (cluster 1) contained 26 strains, which showed that most of the strains are closely related. Included in this cluster, however, are the type strains of Av. volantium (CCUG 3713T), Av. gallinarum (CCUG 12931T), Av. endocarditidis (CCUG 5286T) and Av. avium (CCUG 12833T) as well as the reference strain for Avibacterium species A (CCUG 18782), suggesting that they are all one species. Two subclusters (clusters 1A and 1B) were observed branching at a distance level just below 400. Cluster 1A included the type strains of Av. gallinarum (CCUG 12391T) and Av. endocarditidis (CCUG 5286T). Cluster 1B included the type strain of Av. volantium (CCUG 3713T), Av. avium (CCUG 12833T) and the reference strain of Avibacterium species A (CCUG 18782).
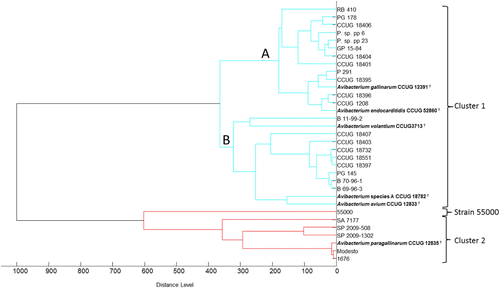
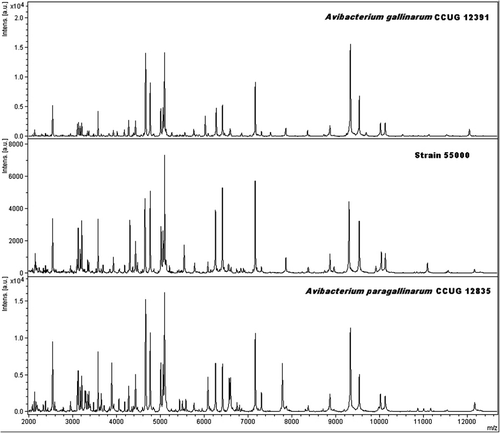
The strain 55000 remained separately on its own at a distance level between 600 and 700. Finally, the type strain of Av. paragallinarum (CCUG 12835T) and two reference strains of serovar B (1676) and serovar C (Modesto) grouped closely together. Cluster 2 also included strains SP2009-508, SP2009-1302 and a reference strain of Av. paragallinarum biovar 2 (SA 7177), which branched from Av. paragallinarum type strain at a distance level above 300. Reference spectra for the different clusters are shown in .
To improve the Avibacterium species identification by MALDI/Biotyper, the six reference strains () were incorporated in the Bruker Biotyper reference database version 3 and identification was again performed. All of the strains were identified at the species level except for strain 55000, which was identified only to the genus level, hence the score value of 1.988 (). Normally, to discriminate two different species, a best score value above 2 for the first species and a second best match below 2 for the second species, which belong to the same genus, have to be achieved (). Therefore, strain 55000 belongs to the Avibacterium genus but to none of the species already known. The organism's best and second best match values () confirm the relationship results observed in the MSP dendrogram. Furthermore, best and second best match values gained for the five different species—Av. volantium (CCUG 3713T), Av. gallinarum (CCUG 12931T), Av. endocarditidis (CCUG 5286T), Av. avium (CCUG 12833T) and Avibacterium species A (CCUG 18782)–demonstrated that they cannot be identified as different species.
Discussion
Previous studies indicate that a reclassification of the genus Avibacterium is required due to severe problems regarding the identification of isolates to species level (Christensen et al., Citation2007; Blackall & Norskov-Lauritsen, Citation2008; Christensen et al., Citation2009; Bisgaard et al., Citation2012). In the present investigation, MALDI/Biotyper was evaluated for identification of members of the genus Avibacterium. In a former study including some 250 non-avian strains representing 15 genera and more than 40 species and subspecies of Pasteurellaceae, a high discrimination at the genus and species level was observed (Kuhnert et al., Citation2012). In the present study, a well-defined strain collection of avian Avibacterium species was investigated. All strains yielded spectra of good and reproducible quality. Although the Biotyper 3.0 reference database includes spectra from type strains of Av. gallinarum, Av. endocarditidis, Av. volantium and Av. avium, identification of the strains investigated was limited to the genus level, with the exception of six strains identified as Av. endocarditidis and two strains that remained undiagnosed. In addition, the reference/type strains of Av. gallinarum, Av. avium and Av. volantium were identified as Av. endocarditidis. The quality of spectra used in MALDI-TOF MS can be influenced by several factors; for example, culture media, culture conditions, and storage conditions (Alispahic et al., Citation2010, Citation2011). In addition, Kuhnert et al. (Citation2012) demonstrated that scores of field strains of Mannheimia tested were always higher with newly generated reference spectra in comparison with those contained in the commercial Biotyper 3.0 database. These observations might explain the present deviations observed for type strains. Furthermore, Van Veen et al. (Citation2010) found that misidentification or poor identification by MALDI-TOF MS was most probably associated with an insufficient number of reference strains available in the database. In addition, problems in resolving closely related species by MALDI-TOF MS were reported by Duskova et al. (Citation2012) and Kuhnert et al. (Citation2012).
According to Sauer et al. (Citation2008) clusters of strains with distance levels less than 500 have been described as reliably classified species. In addition, Kuhnert et al. (Citation2012) stated that the dendrogram derived from similarity matrices based on MSP clearly showed that strains of the same species formed a well-defined narrow cluster with a distance level less than 100. The different distance levels observed are probably taxon dependent.
In the present study, MALDI-TOF MS revealed that the type and reference strains of Av. volantium, Av. gallinarum, Av. endocarditidis, Av. avium and Avibacterium species A clustered together with 26 field isolates in cluster 1, at a distance level below 400. Comparison of these results with the sequence types determined by Bisgaard et al. (Citation2012) revealed no obvious links. Two sequence types (1, 22) even had strains located in both subclusters 1A and 1B. Similarly, strains clustering together in the concatenated tree based upon rpoB, pgi, recN, sodA and infB (Bisgaard et al., Citation2012) were located in both subclusters. In the present investigation, the type strains of Av. gallinarum and Av. endocarditidis clustered together at a distance level of less than 100, indicating that they could belong to the same species (Kuhnert et al., Citation2012). In the concatenated tree based upon rpoB, pgi, recN, sodA and infB sequences, however, these strains clustered separately. In addition, genome similarity as demonstrated by the average nucleotide identity only showed 70% similarity (Bisgaard et al., Citation2012). The diversity and lack of cohesion might be due to the fact that members of Avibacterium have not fully diverged to represent genuine species (Bisgaard et al., Citation2012), still having traits from another species as described previously for other bacteria (Retchless & Lawrence, Citation2007). Also, the fact that the sequence types are based on the analysis of several selected genes and MALDI-TOF MS bases on protein profiles of a cell might contribute to such differences. The three reference strains of serovars A, B and C of Av. paragallinarum belong to biovar 1, in cluster 2, demonstrating a distance level of less than 100. According to MALDI-TOF MS, these strains are clearly identifiable as a unique species in agreement with the results of Kuhnert et al. (Citation2012) and Bisgaard et al. (Citation2012). However, the low differences observed between serovars do not seem to provide a possibility for identification of individual serovars by MALDI-TOF MS; ideally, multiple strains belonging to the same serovar are needed to confirm this statement.
The unique position of biovar 2 of Av. paragallinarum (SA 7177) was surprising. This strain clustered together with two biovar 1 strains (SP2009-1302 and SP2009-508) at a distance level less than 400. Strain SP2009-508 was previously found to differ compared with the type strain of Av. paragallinarum in sequences of rpoB, recN, infB and pgi, whereas SP2009-1302 showed high similarity to Av. paragallinarum in these genes. Both strains were detected with the Av. paragallinarum-specific polymerase chain reaction of Chen et al. (Citation1996). This group of bacteria clustered at a depth of more than 400 with the type Av. paragallinarum type strain. This finding is in agreement with the results obtained by polymerase chain reaction that identified these isolates as Av. paragallinarum (Bisgaard et al., Citation2012). However, genotypic data revealed considerable differences between the type strain of Av. paragallinarum and SP2009-1302 (Bisgaard et al., Citation2012). Strain 55000, separating on its own above a distance level of 600 to the other Av. paragallinarum strains, indicating the existence of a different species. This strain was isolated from a pheasant showing sinusitis and tested negative for Av. paragallinarum by polymerase chain reaction (Bisgaard et al., Citation2012). Except for genuine Av. paragallinarum represented by reference strains of the serovars A, B and C in the current study, atypical strains of Av. paragallinarum like V-factor independent strains such as SP2009-508 and SP2009-1302 will only be identified if included in the database. The same is valid for isolates associated with endocarditis in adult broiler breeders.
Kuhnert et al. (Citation2012) stated that MALDI-TOF MS may be used as a screening method for species not properly recognized to extend our knowledge on the significance of new taxa. The present investigation clearly confirmed this statement and the conclusion of Bisgaard et al. (Citation2012) that the species Av. volantium, Av. gallinarum, Av. endocarditidis, Av. avium and Avibacterium species A might represent incipient species. However, in this strain collection the Av. avium species and the Av. volantium species contain just one strain belonging to each species, which could limit the conclusions that can be reached on these species. Finally, MALDI-TOF MS was shown to represent a fast and reliable method for identification and separation of the Av. paragallinarum complex. The Bruker reference database is continually updated; consequently, during the manuscript review the Av. endocarditidis DSM 18224T strain was removed.
The presented data are in agreement with the findings of Bisgaard et al. (Citation2012) suggesting a re-organization of the taxonomy of the genus Avibacterium. For this purpose, however, further taxonomic investigations including whole genome comparisons are needed.
Acknowledgement
Excellent technical assistance was contributed by Delfina Jandreski-Cvetkovic.
Funding
This work was partly done within the CEPO (Centre of Excellence for Poultry) project, funded by the European Regional Development Fund, Cross-border Cooperation Programme Austria-Hungary 2007–2013.
Additional information
Funding
References
- Alispahic, M., Hummel, K., Jandreski-Cvetkovic, D., Nobauer, K., Razzazi-Fazeli, E., Hess, M. & Hess, C. (2010). Species-specific identification and differentiation of Arcobacter, Helicobacter and Campylobacter by full-spectral matrix-associated laser desorption/ionization time of flight mass spectrometry analysis. Journal of Medical Microbiology, 59, 295–301. 10.1099/jmm.0.016576-0
- Alispahic, M., Christensen, H., Hess, C., Razzazi-Fazeli, E., Bisgaard, M. & Hess, M. (2011). Identification of Gallibacterium species by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry evaluated by multilocus sequence analysis. International Journal of Medical Microbiology, 301, 513–522. 10.1016/j.ijmm.2011.03.001
- Bisgaard, M., Norkov-Lauritson, N., De Wit, S.J., Hess, C. & Christensen, H. (2012). Multilocus sequence phylogenetic analysis of Avibacterium. Microbiology, 158, 993–1004. 10.1099/mic.0.054429-0
- Bisgaard, M., Christensen, J.P., Bojesen, A.M. & Christensen, H. (2007). Avibacterium endocarditidis sp. nov., isolated from valvular endocarditis in chickens. International Journal of Systematic and Evolutionary Microbiology, 57, 1729–1734. 10.1099/ijs.0.64879-0
- Blackall, P.J. (1988). Biochemical properties of catalase-positive avian haemophili. Journal of General Microbiology, 134, 2801–2805.
- Blackall, P.J., Christensen, H., Beckenham, T., Blackall, L.L. & Bisgaard, M. (2005). Reclassification of Pasteurella galllinarum, [Haemophilus] paragallinarum, Pasteurella avium and Pasteurella volantium as Avibacterium gallinarum gen. nov., comb. nov., Avibacterium paragallinarum comb. nov., Avibacterium avium comb. nov. and Avibacterium volantium comb. nov. International Journal of Systematic and Evolutionary Microbiology, 55, 353–362. 10.1099/ijs.0.63357-0
- Blackall, P.J. & Norskov-Lauritsen, N. (2008). Pasteurellaceae – the view from the diagnostic laboratory. In P. Kuhnert & H. Christensen. (Eds.). Pasteurellaceae: Biology, Genomics and Molecular Aspects (pp. 227–259). Norfolk: Caister Academic Press.
- Chen, X., Miflin, J.K., Zhang, P. & Blackall, P.J. (1996) Development and application of DNA probes and PCR tests for Haemophilus paragallinarum. Avian Diseases, 40, 398–407. 10.2307/1592238
- Christensen, H., Kuhnert, P., Busse, H.J., Frederiksen, W.C. & Bisgaard, M. (2007). Proposed minimal standards for the description of genera, species and subspecies of the Pasteurellaceae. International Journal of Systematic and Evolutionary Microbiology, 57, 166–178. 10.1099/ijs.0.64838-0
- Christensen, H., Blackall, P.J. & Bisgaard, M. (2009). Phylogenetic relationships of unclassified, satellitic Pasteurellaceae obtained from different species of birds as demonstrated by 16S rRNA gene sequence comparison. Research in Microbiology, 160, 315–321. 10.1016/j.resmic.2009.05.006
- Duskova, M., Sedo, O., Ksicova, K., Zdrahal, Z. & Karpinskova, R. (2012). Identification of lactobacilli isolated from food by genotypic methods and MALDI-TOF MS. International Journal of Food Microbiology, 159, 107–114. 10.1016/j.ijfoodmicro.2012.07.029
- Freiwald, A. & Sauer, S. (2009). Phylogenetic classification and identification of bacteria by mass spectrometry. Nature Protocols, 4, 732–742. 10.1038/nprot.2009.37
- Kuhnert, P., Bisgaard, M., Korczak, B.M., Schwendener, S., Christensen, H. & Frey, J. (2012). Identification of animal Pasteurellaceae by MALDI-TOF mass spectrometry. Journal of Microbiological Methods, 89, 1–7. 10.1016/j.mimet.2012.02.001
- Mutters, R., Piechulla, K., Hinz, K.H. & Mannheim, W. (1985). Pasteurella avium (Hinz and Kunjara 1977) comb. nov. and Pasteurella volantium sp. nov. International Journal of Systematic Bacteriology, 35, 5–9. 10.1099/00207713-35-1-5
- Retchless, A.C. & Lawrence, J.G. (2007). Temporal fragmentation of speciation in bacteria. Science, 317, 1093–1096. 10.1126/science.1144876
- Sauer, S., Freiwald, A., Maier, T., Kube, M., Reinhardt, R., Kostrzewa, M. & Geider, K. (2008). Classification and identification of bacteria by mass spectrometry and computational analysis. PLOS ONE 3, 2843. 10.1371/journal.pone.0002843
- Sauer, S. & Kliem, M. (2010). Mass spectrometry tools for the classification and identification of bacteria. Nature Reviews Microbiology, 8, 74–82. 10.1038/nrmicro2243
- Seng, P., Drangcourt, M., Gouriet, F., La Scola, B., Fournier, P.E., Rolain, J.M. & Raoult, D. (2009). Ongoing revolution in bacteriology: routine identification of bacteria by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Clinical Infectious Diseases, 49, 543–551. 10.1086/600885
- Van Veen, S.Q., Claas, E.C.J. & Kuijper, E.J. (2010). High-throughput identification of bacteria and yeast by Matrix-Assisted Laser Desorption Ionization-Time of Flight Mass Spectrometry in conventional medical microbiology laboratories. Journal of Clinical Microbiology, 48, 900–907. 10.1128/JCM.02071-09
