Abstract
The highly pathogenic avian influenza virus (HPAIV) subtype H5N1 threatens animal and human health worldwide. Susceptibility of pigeons to HPAIV (H5N1) and their role in avian influenza virus transmission to domestic birds and humans remain questionable. In this study, an outbreak in domestic pigeons (1 to 18 months old) with 50% mortality was investigated. Pigeons exhibited nervous manifestations and greenish diarrhoea. Necropsy of the naturally infected pigeons revealed congestion of the internal organs, particularly the lungs and brain. The HPAIV subtype H5N1 designated A/Pigeon/Egypt/SHAH-5803/2011 was isolated from a 40-day-old pigeon. Sequencing of the haemagglutinin gene showed it to be closely related to viruses in group 2.2.1/C. Intravenous inoculation of the isolate in chickens induced 100% mortality within 2 days post inoculation and the intravenous pathogenicity index was 2.7. Virus pathogenicity and transmissibility was determined experimentally in 6-week-old domestic pigeons. Thirty per cent of pigeons inoculated oronasally with 106 median embryo infective dose showed congested beak, conjunctivitis, depression, and greenish diarrhoea. A mortality rate of 10% was recorded preceded by severe neurologic signs consisting of torticollis, incoordination, tremors, and wing paralysis. Pathological examination revealed a friable brain tissue and congested meningeal blood vessels. The lungs appeared oedematous and severely haemorrhagic. Subepicardial and petechial haemorrhages on the coronary fat were observed. Both infected and contact pigeons shed virus via the oropharynx and cloaca. To our knowledge, this is the first description and characterization of HPAIV in naturally infected pigeons in Egypt. Our findings reveal that pigeons can indeed be susceptible to H5N1 HPAIVs and could be a source of infection to other birds and humans.
Introduction
Influenza A viruses belong to the family Orthomyxoviridae and have an enveloped, segmented, single-stranded, and negative-sense RNA genome. Influenza A viruses of avian origin can be categorized into subtypes based on the two surface glycoproteins, 16 different types of haemagglutinin (HA) and nine different types of neuraminidase (Fouchier et al., Citation2005). Avian influenza viruses (AIVs) are classified into two groups, highly pathogenic and low pathogenic, based on the severity of the disease they cause in chickens (Swayne & Suarez, Citation2000). Highly pathogenic avian influenza viruses (HPAIVs), which code for a furin-sensitive cleavage site in their HA protein, are capable of inducing systemic infections with dramatic losses in the poultry industry, particularly chickens and turkeys (Alexander, Citation1993).
Highly pathogenic avian influenza, an important transboundary disease, poses a threat to both animal and public health. Controversial reports exist about the pigeon's susceptibility to HPAIV. Their role as a link between wild and domestic birds as well as the transmission and spread of HPAIV during epizootics over long distances is not yet clear (Kaleta & Hönicke, Citation2004). Although Asian-lineage H5N1 HPAIV has been isolated from pigeons (Ellis et al., Citation2004), the existing evidence indicates that pigeons are only minimally susceptible or resistant to different subtypes of AIV infection and do not serve as an efficient transmission host, even in the presence of immune dysfunction (Fang et al., Citation2006) such as immunosuppression by cyclophosphamide (Panigrahy et al., Citation1996; Perkins & Swayne, Citation2002; Kaleta & Hönicke, Citation2004; Liu et al., Citation2007; Smietanka et al., Citation2011). Pigeons probably played a minimal epidemiologic role in the perpetuation of the H5N1 Hong Kong-origin influenza viruses (Perkins & Swayne, Citation2002). Other experimental studies have demonstrated that pigeons are susceptible to highly pathogenic influenza viruses (Klopfleisch et al., Citation2006; Werner et al., Citation2007; Jia et al., Citation2008; Smietanka et al., Citation2011) and could be a source of infection for other animals (Songserm et al., Citation2006).
The susceptibility of pigeons to H5N1 virus and their role in AIV transmission to domestic birds and humans remain questionable. Here we report for the first time in Egypt a case of natural infection with HPAIV H5N1 in pigeons. The characterization, pathogenicity and transmissibility of the field AIV isolated from a pigeon were investigated.
Materials and Methods
Samples and virus isolation
In March 2011, two dead pigeon squabs (40 days old) were submitted to the Clinic of Avian and Rabbit Medicine Department, Faculty of Veterinary Medicine, Zagazig University, Egypt. According to the owner, the pigeons exhibited greenish diarrhoea, nervous manifestations and sudden deaths with a mortality rate up to 50% within 10 days. The flock consisted of 50 non-vaccinated birds of variable ages (1 to 18 months) reared in a backyard free-range system. The pigeons were necropsied and pooled respiratory samples (lung and trachea) were collected and injected into 10-day-old specific pathogen free embryonated chicken eggs via the allantoic route according to recommendations of the OIE (Citation2012). The allantoic was collected fluid from dead embryos and screened by rapid haemagglutination test. The infected allantoic fluid was then tested for the presence of AIV by reverse transcriptase-polymerase chain reaction (RT-PCR) and further sequenced. Newcastle disease virus infection was excluded in HA-positive allantoic fluids by negative results of both the Newcastle Disease Virus Antigen Rapid Test (Shenzhen Combined Biotech Co. Ltd, Shenzhen, China) and RT-PCR as described by Pang et al. (Citation2002).
RNA extraction and reverse transcriptase-polymerase chain reaction
Viral RNA was extracted from infected allantoic fluid using the Gene JET™ RNA purification kit (catalogue number K0731; Fermentas, EU, Vantaa, Finland) according to the manufacturer's instructions. Five microlitres of extracted RNA was reverse transcribed to cDNA using the RevertAid™ H Minus First Strand cDNA synthesis kit (catalogue number K1611; Fermentas, EU) following the manufacturer's instructions. The PCR was performed with a set of primers specific for H5 and N1 gene segments as described elsewhere (Njouom et al., Citation2008).
Viral sequencing and phylogenetic analysis
For characterization of the isolated virus, the HA gene was analysed by PCR using a set of HA gene-specific primers (Njouom et al., Citation2008). The PCR product was gel purified using the Gene JET™ Gel Extraction Kit (catalogue number K0691; Fermentas, EU) as recommended by the manufacturer and was sequenced on both strands using the amplification primers (Solgent Co. Ltd, Daedeokdaero Yuseong-gu Daejeon, Korea). The HA subtype was further identified by nucleotide BLAST (http://www.ncbi.nlm.nih.gov/BLAST) and submitted to GenBank. The obtained nucleotide sequence was aligned with other HA gene sequences available in GenBank and comparative alignment was performed by the ClustalW method using the MegAlign module of DNAStar software (Lasergene version 7.2; DNASTAR, Madison, Wisconsin, USA). Phylogenetic analysis was carried out on the HA gene region encompassing 1594 nucleotides. Forty-eight reference sequences retrieved from the GenBank database were used in this study to understand the relationship of the isolated virus with other co-circulating H5N1 viruses from birds and mammals. Phylogenetic comparison of the aligned sequences was generated using the neighbour-joining method employing the Kimura two-parameter correction in MEGA version 5 (www.megasoftware.net). The reliability of internal branches was assessed by 1000 bootstrap replications and the p-distance substitution model. The H5 numbering used throughout the study was based on the alignment with A/Goose/Guangdong/1/96 (H5N1) minus the 16 amino acids known as HA signal peptide.
Chicken intravenous pathogenicity test
A pathogenicity test was performed and the intravenous pathogenicity index of the virus in chickens was determined according to the guidelines in the OIE manual (OIE, Citation2012).
Pathogenicity and transmissibility of AIV isolated from naturally infected pigeons
To determine whether the isolated H5N1 virus would produce infection under experimental conditions, a group of 10 healthy 6-week-old domestic pigeons, serologically negative for AIV H5N1 subtype, were inoculated oronasally with 0.1 ml virus fluid containing 106 median embryo infective dose (EID50). Six pigeons were added as sentinels into the pigeons’ aviary to detect transmissibility. The sentinel pigeons had direct contact with the inoculated pigeons’ faeces and a common source of drinking water. Additionally, three sham-inoculated control pigeons were housed separately from the other birds as a negative control group. All pigeons were monitored two or three times daily for a total of 14 days and clinical signs were recorded. Oropharyngeal and cloacal swabs were taken from pigeons at 3, 5 and 7 days post infection (d.p.i.) for virus re-isolation and titration. The experimental studies were permitted by the Committee for Community Service and Environment Development, Zagazig University, Egypt.
Pathology
Pigeons were euthanized at 14 d.p.i., were necropsied and internal organs were sampled for virus re-isolation. Selected organs were fixed in 10% neutral buffered formalin, routinely processed and embedded in paraffin (Allen, Citation1994). Paraffin blocks were sectioned in duplicate at 5 µm and routinely stained by haematoxylin and eosin. Consecutive sections were stained by immunohistochemistry to determine the AIV antigen distribution in tissues. A monoclonal antibody against the influenza A virus nucleoprotein (ATCC HB-65 influenza A hybridoma cell line) was used as the primary antibody. Specific antigen–antibody reactions were visualized by 3,3′-diaminobenzidine tetrahydrochloride treatment using the Dako EnVision system (DakoCytomation Inc. Carpinteria, CA, USA) and counterstained with haematoxylin. Positive results appeared as brown precipitate localized at the site of binding (Perkins & Swayne, Citation2001).
Virus re-isolation and identification
The collected tissue and swab samples were prepared and inoculated into 10-day-old specific pathogen free embryonated chicken eggs via the allantoic cavity for virus isolation (OIE, Citation2012). Infectivity titres were calculated by the method of Reed & Muench (Citation1938). Viral RNA was extracted and RT-PCR was performed to detect the presence of AIV as described previously. Newcastle disease virus infection was excluded in HA-positive allantoic fluids by a negative RT-PCR result as described in Pang et al. (Citation2002).
Nucleotide sequence accession number
The nucleotide sequence obtained from this study is available at GenBank under accession number JX965419.
Results
Detection and isolation of avian influenza virus
Necropsied naturally infected pigeons showed congestion of the internal organs, particularly the lungs and brain. The virus isolated in this study, designated A/Pigeon/Egypt/SHAH-5803/2011 and determined as H5N1 by RT-PCR, caused embryo deaths within 3 d.p.i. and showed a titre of 1/32 HA units.
Molecular and biological features of pigeon AIV
Molecular characterization of the pigeon AIV isolate was performed by HA gene sequencing and nucleotide BLAST analysis. The Egyptian pigeon strain possessed multi-basic amino acids at the connecting peptide between HA1 and HA2 (PQGERRRKKR/GLF), typical for HPAIV of clade 2.2, as well as glutamine and glycine (Q222–G224) at the receptor binding site and the presence of amino acid D387 (H5 HA numbering), characteristic to sub-clade 2.2.1. The HA gene of the investigated pigeon isolate revealed that it belonged to the 5J lineage according to the Influenza A Virus Genotype Tool (Lu et al., Citation2007).
Virulence of A/Pigeon/Egypt/SHAH-5803/2011 was evaluated through intravenous challenge of chickens. Mortality in chickens was 100% within 48 h. Chickens showed signs of depression, anorexia, oedema and haemorrhage in the comb and shanks, and demonstrated an intravenous pathogenicity index of 2.7.
Phylogenetic analysis
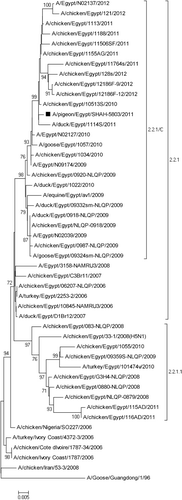
The pigeon HA gene sequence had a nucleotide similarity of 95.2 to 99.6% and 97.6 to 99.4% with Egyptian AIV H5N1 subtypes isolated from birds and mammals respectively. The HA gene of the pigeon AIV was phylogenetically closely related to viruses in the 2.2.1/C group isolated from backyard chickens, ducks and human (). A deletion within the receptor binding site at position 129S and an additional seven amino acid substitutions (D43N, S120D, I151T, D154N, N155A, R162K and G272S) of the viral H5 protein were found in comparison with H5N1 virus introduced into Egypt in 2006 (Index/2006).
Experimental infection
Pathology
Infected pigeons (3/10 birds) showed nasal congestion, conjunctivitis and slight depression at 2 d.p.i. At 4 d.p.i., severe neurologic signs consisting of torticollis (), incoordination, tremors and wing paralysis followed by death at 5 d.p.i. were seen (1/10 bird). Greenish diarrhoea was seen at 7 d.p.i. (3/10 birds). No clinical signs were observed in the sham-inoculated control pigeons.
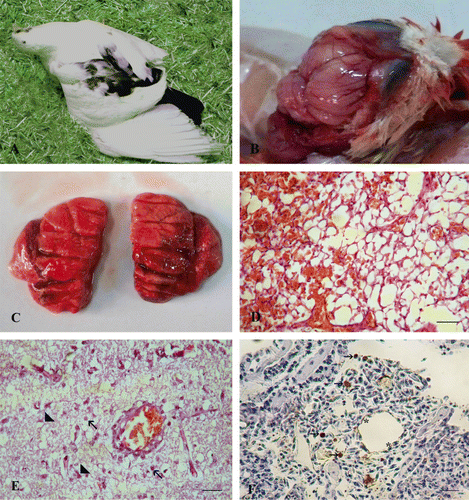
Gross examination revealed a soft and friable brain tissue with congested meningeal blood vessels (). The lungs appeared oedematous and severely haemorrhagic (infected pigeon at 5 d.p.i.) (). Subepicardial and petechial haemorrhages on the coronary fat were observed. Histologically, the liver showed focal aggregations of leukocytes in the portal areas and congestion of the blood vessels. Pulmonary oedema, congestion of pulmonary blood vessels and occlusion of the alveolar spaces with erythrocytes were observed (). The brain suffered from vacuolation, spongiosis, degenerated neurons, microglial cell infiltrations and perivascular lymphocytic cuffing as well as congested blood vessels of the meninges (). Sporadic moderate positive staining of the pneumocytes, interstitial tissue and few intravascular mononuclear cells of the lungs was observed (). Viral antigens were detected in the brain and intestinal tract as well. The sham-inoculated control pigeons showed no lesions and remained negative on immunostaining.
Virus excretion and transmission kinetics
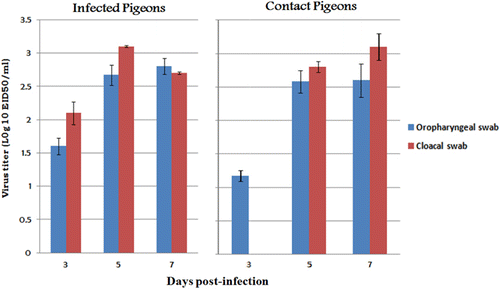
Systemic infection through 5 d.p.i. was confirmed in the infected pigeons. The virus titres ranged from 103.75 to 104.5 EID50/g. Moreover, the virus was isolated from the brain and intestinal tissues of the sentinel pigeons at 14 d.p.i. Oropharyngeal and cloacal swabs were taken from pigeons to detect virus shedding. The virus was re-isolated from collected swabs, with titres ranging from 101.6 to 103.16 EID50/ml and from 101.17 to 103.1 EID50/ml in infected and sentinel pigeons respectively (). Swabs and tissue samples of sham-inoculated control pigeons were negative.
Discussion
Avian influenza H5 subtype viruses have caused destructive outbreaks in domestic poultry and several human infections, raising the fear of a pandemic. In the present study, H5N1 virus was isolated and identified from respiratory samples of naturally infected pigeons in Sharkia Province, Egypt during 2011. Based on the phylogenetic analysis of the HA gene sequences, the virus clusters into group 2.2.1/C isolated from backyard birds, the live bird market and human (Abdelwhab et al., Citation2010; Hafez et al., Citation2010). Since pigeons commonly reside with humans on farms in Egypt, their role as potential carriers for H5N1 virus should not be neglected. Being the most abundant birds on backyard poultry farms, pigeons can become H5N1 virus-infected during feeding when intermingling freely with domestic and aquatic birds.
A major molecular determinant for pathogenicity of H5 and H7 viruses is the amino acid sequence specifying the proteolytic cleavage site of HA. The HA mediates the attachment of the virus to sialic-acid-containing receptors and the fusion of the virus envelope with the endosomal membrane (Rott et al., Citation1995; Katz et al., Citation2000). The HA molecule of the pigeon isolate contains amino acid residues 222Q and 224G at the receptor-binding site, which determines the binding of the HA molecule to avian-specific sialic acid α-2,3-galactose receptors (Matrosovich et al., Citation1999). Previously, it was proposed that isolates having isoleucine at residue 151 of the HA1 protein might be involved in different receptor binding (Claas et al., Citation1998). Recently, it was shown that deletion of RBD 129S combined with I151T increased α2–6 SA binding (Watanabe et al., Citation2011). It was observed that the pigeon strain isolated in this study has these substitutions. In addition, specific amino acid positions Y94, S132, W149, H179, E186, K189, L190, E212, P217, K218, G221, Q222, S223 and G224 (H5 influenza numbering), implicated in receptor specificity (Stevens et al., Citation2006), were further investigated. Interestingly, the substitutions at positions 94 (Y to N), 189 (K to R), 212 (E to K) and 217 (P to S) that were found in the examined pigeon isolate have also been observed in the HA gene sequence of A/equine/Egypt/av1/2009 isolated from donkeys in Egypt (Abdel-Moneim et al., Citation2010).
The present study also demonstrates the ability of an Egyptian-origin HPAIV to replicate in experimentally infected pigeons, inducing 30% morbidity and 10% mortality, and to be transmitted to contact birds. These findings are different from previous research showing that pigeons were mostly resistant and probably played a minimal epidemiological role in the dissemination of the H5N1 Hong Kong-origin influenza viruses (Perkins & Swayne, Citation2002). Similar results were reported by Klopfleisch et al., (Citation2006) in pigeons experimentally infected with a high infective dose (108 EID50) of HPAIV (A/chicken/Indonesia/2003), indicating that different H5N1 viruses have distinctive biological properties. In this study, the aggressive outbreak recorded in the naturally infected pigeons could not be reproduced experimentally. This may also be attributed to co-infection with other pathogens, bad hygiene and/or stress of flying. Thus, our study further indicates that H5N1 HPAIVs have established infection and pathogenicity among various aquatic and wild birds, suggesting that the viruses are continuously evolving compared with previously endemic HPAIVs (Ellis et al., Citation2004; Chen et al., Citation2006).
In experiments with HPAIV H5N1 in gallinaceous birds, the virus produced a fulminating and rapidly fatal systemic disease (Perkins & Swayne, Citation2001). The results from experimental infection performed in this study using oronasal inoculation in pigeons were similar to those found in the previous study with regards to systemic replication. The virus reached a titre of 104.5 EID50/g in the brains of experimentally infected pigeons, but only one pigeon showed neurological manifestations and histologically showed severe vacuolization, spongiosis and neuronal degeneration. A pronounced neurotropism of HPAIV A/chicken/Indonesia/2003 (H5N1) for the cerebrum and brainstem of pigeons was recorded (Klopfleisch et al., Citation2006). Moreover, our data showed that pigeons excreted infectious viruses via the oral or cloacal routes. In contrast to a previous report (Werner et al., Citation2007), the virus was able to trigger an infection in contact non-inoculated pigeons. Strikingly, 70% of the infected pigeons had considerable virus titre in their tissues and excreta without clinical signs and can thus pass the virus silently and act as a reservoir.
In conclusion, this study is the first to report the entire HA gene sequence of H5N1 virus isolated from naturally infected pigeons in Egypt. The results of this study revealed that H5N1 HPAIVs are continually evolving and changing their pathogenicity to be able to replicate and shed from infected pigeons, consequently increasing the risk of transmission to other avian species and humans and emphasizing the importance of full genome sequencing of pigeon H5N1 isolates. Knowledge about the circulating genomes may increase understanding of the spread and mechanisms of virulence and pathogenicity of H5N1 viruses to poultry and humans. Further surveys, particularly continuing surveillance of pigeons, are necessary.
Acknowledgements
The authors would like to thank the Department of Veterinary and Biomedical Sciences, Animal Disease Research and Diagnostic Laboratory, South Dakota State University, Brookings, South Dakota, USA for supporting the material and equipment used in immunohistochemistry staining, and Prof. Dr Christopher C.L. Chase, South Dakota State University for his valuable scientific revision of the manuscript.
References
- Abdel-Moneim, A.S., Abdel-Ghany, A.E. & Shany, S.A. (2010). Isolation and characterization of highly pathogenic avian influenza virus subtype H5N1 from donkeys. Journal of Biomedical Science, 17, 25. 10.1186/1423-0127-17-25
- Abdelwhab, E.M., Selim, A.A., Arafa, A., Galal, S., Kilany, W.H., Hassan, M.K., Aly, M.M. & Hafez, M.H. (2010). Circulation of avian influenza H5N1 in live bird markets in Egypt. Avian Diseases, 54, 911–914. 10.1637/9099-100809-RESNOTE.1
- Alexander, D.J. (1993). Virus infection of birds. In J.B. McFerran & M.S. McNulty (Eds.). Virus Infection of Birds (pp. 287–316). London: Elsevier Science.
- Allen, T.C. (1994). Hematoxylin and eosin. In E.B. Prophet, B. Mills, J.B. Arrington & L.H. Sobin (Eds.). Laboratory Methods in Histotechnology (pp. 53–58). Washington: American Registry of Pathology.
- Chen, H., Li, Y., Li, Z., Shi, J., Shinya, K., Deng, G., Qi, Q., Tian, G., Fan, S., Zhao, H., Sun, Y. & Kawaoka, Y. (2006). Properties and dissemination of H5N1 viruses isolated during an influenza outbreak in migratory waterfowl in western China. Journal of Virology, 80, 5976–5983. 10.1128/JVI.00110-06
- Claas, E.C., Osterhaus, A.D., van Beek, R., De Jong, J.C., Rimmelzwaan, G.F., Senne, D.A., Krauss, S., Shortridge, K.F. & Webster, R.G. (1998). Human influenza A H5N1 virus related to a highly pathogenic avian influenza virus. Lancet, 351, 472–477. 10.1016/S0140-6736(97)11212-0
- Ellis, T.M., Bousfield, R.B., Bissett, L.A., Dyrting, K.C., Luk, G.S., Tsim, S.T., Sturm-Ramirez, K., Webster, R.G., Guan, Y. & Peiris, J.S.M. (2004). Investigation of outbreaks of highly pathogenic H5N1 avian influenza in waterfowl and wild birds in Hong Kong in late 2002. Avian Pathology, 33, 492–505. 10.1080/03079450400003601
- Fang, T.H., Lien, Y.Y., Cheng, M.C. & Tsai, H.J. (2006). Resistance of immune-suppressed pigeons to subtypes H5N2 and H6N1 low pathogenic avian influenza virus. Avian Diseases, 50, 269–272. 10.1637/7437-090905R.1
- Fouchier, R.A., Munster, V., Wallensten, A., Bestebroer, T.M., Herfst, S., Smith, D., Rimmelzwaan, G.F., Olsen, B. & Osterhaus, A.D. (2005). Characterization of a novel influenza A virus hemagglutinin subtype (H16) obtained from black-headed gulls. Journal of Virology, 79, 2814–2822. 10.1128/JVI.79.5.2814-2822.2005
- Hafez, M.H., Arafa, A., Abdelwhab, E.M., Selim, A., Khoulosy, S.G., Hassan, M.K. & Aly M.M. (2010). Avian influenza H5N1 virus infections in vaccinated commercial and backyard poultry in Egypt. Poultry Science, 89, 1609–1613. 10.3382/ps.2010-00708
- Jia, B., Shi, J., Li, Y., Shinya, K., Muramoto, Y., Zeng, X., Tian G., Kawaoka Y & Chen, H. (2008). Pathogenicity of Chinese H5N1 highly pathogenic avian influenza viruses in pigeons. Archives of Virology, 153, 1821–1826. 10.1007/s00705-008-0193-8
- Kaleta, E.F. & Hönicke. A. (2004). Review of the literature on avian influenza A viruses in pigeons and experimental studies on the susceptibility of domestic pigeons to influenza A viruses of the haemagglutinin subtype H7. DTW Deutsche tierärztliche Wochenschrift, 111, 467–472.
- Katz, J.M., Lu, X., Tumpey, T.M., Smith, C.B., Shaw, M.W. & Subbarao, K. (2000). Molecular correlates of influenza A H5N1 virus pathogenesis in mice. Journal of Virology, 74, 10807–10810. 10.1128/JVI.74.22.10807-10810.2000
- Klopfleisch, R., Werner, O., Mundt, E., Harder, T. & Teifke, J. (2006). Neurotropism of highly pathogenic avian influenza virus A/chicken/Indonesia/2003 (H5N1) in experimentally infected pigeons (Columba livia f. domestica). Veterinary Pathology, 43, 463–470. 10.1354/vp.43-4-463
- Liu, Y., Zhou, J., Yang, H., Yao, W., Bu, W., Yang, B., Song, W., Meng, Y., Lin, J., Han, C., Zhu, J., Ma, Z., Zhao, J. & Wang, X. (2007). Susceptibility and transmissibility of pigeons to Asian lineage highly pathogenic avian influenza virus subtype H5N1. Avian Pathology, 36, 461–465. 10.1080/03079450701639335
- Lu, G., Rowley, T., Garten, R. & Donis, R.O. (2007). FluGenome: a web tool for genotyping influenza A virus. Nucleic Acids Research, 35, 275–279. 10.1093/nar/gkm365
- Matrosovich, M., Zhou, N., Kawaoka, Y. & Webster, R. (1999). The surface glycoproteins of H5 influenza viruses isolated from humans, chickens, and wild aquatic birds have distinguishable properties. Journal of Virology, 73, 1146–1155.
- Njouom, R., Aubin, J.T., Bella, A.L., Demsa, B.M., Rouquet, P., Gake, B., Ngangnou, A., Foupouapouognigni, Y., Van Der Werf, S., Rocourt, J. & Rousset, D. (2008). Highly pathogenic avian influenza virus subtype H5N1 in ducks in the Northern part of Cameroon. Veterinary Microbiology, 130, 380–384. 10.1016/j.vetmic.2008.02.006
- OIE. (2012). Manual of Diagnostic Tests and Vaccines for Terrestrial Animals (Chapter 2.3.4). http://www.oie.int/en/international-standard-setting/terrestrial-manual/access-online/ accessed on 25 May 2013.
- Pang, Y., Wang, H., Girshick, T., Xie, Z. & Khan, M.I. (2002). Development and application of a multiplex polymerase chain reaction for avian respiratory agents. Avian Diseases, 46, 691–699. 10.1637/0005-2086(2002)046[0691:DAAOAM]2.0.CO;2
- Panigrahy, B., Senne, D.A., Pedersen, J.C., Shafer, A.L., & Pearson, J.E. (1996). Susceptibility of pigeons to avian influenza. Avian Diseases, 40, 600–604. 10.2307/1592270
- Perkins, L.E.L. & Swayne, D.E. (2001). Pathobiology of A/chicken/Hong Kong/220/97 (H5N1) avian influenza virus in seven gallinaceous species. Veterinary Pathology, 38, 149–164. 10.1354/vp.38-2-149
- Perkins, L.E.L. & Swayne, D.E. (2002). Pathogenicity of a Hong Kong origin H5N1 highly pathogenic avian influenza virus for emus, geese, ducks, and pigeons. Avian Diseases, 46, 53–63. 10.1637/0005-2086(2002)046[0053:POAHKO]2.0.CO;2
- Reed, L. & Muench, H. (1938). A simple method of estimating fifty percent endpoints. American Journal of Hygiene, 27, 493–497.
- Rott, R., Klenk, H.D., Nagai, Y. & Tashiro, M. (1995). Influenza viruses, cell enzymes, and pathogenicity. American Journal of Respiratory and Critical Care Medicine, 152, S16–S19. 10.1164/ajrccm/152.4_Pt_2.S16
- Smietanka, K., Minta, Z., Wyrostek, K., Józwiak, M., Olszewska, M., Domanska-Blicharz, A.K., Reichert, A.M., Pikuła, A., Habyarimana, A. & van den Berg, T. (2011). Susceptibility of pigeons to Clade 1 and 2.2 High Pathogenicity Avian Influenza H5N1 virus. Avian Diseases, 55, 106–112. 10.1637/9514-090110-ResNote.1
- Songserm, T., Amonsin, A., Jam-on, R., Sae-Heng, N., Meemak, N., Pariyothorn, N., Payungporn, S., Theamboonlers, A. & Poovorawan, Y. (2006). Avian influenza H5N1 in naturally infected domestic cats. Emerging Infectious Diseases, 12, 681–683. 10.3201/eid1204.051396
- Stevens, J., Blixt, O., Tumpey, T.M., Taubenberger, J.K., Paulson, J.C. & Wilson, I.A. (2006). Structure and receptor specificity of the hemagglutinin from an H5N1 influenza virus. Science, 312, 404–410. 10.1126/science.1124513
- Swayne, D.E. & Suarez, D.L. (2000). Highly pathogenic avian influenza. Revue scientifique et technique (International Office of Epizootics), 19, 463–482.
- Watanabe, Y., Ibrahim, M.S., Ellakany, H.F., Kawashita, N., Mizuike, R., Hiramatsu, H., Sriwilaijaroen, N., Takagi, T., Suzuki, Y. & Ikuta, K. (2011). Acquisition of human-type receptor binding specificity by new H5N1 influenza virus sublineages during their emergence in birds in Egypt. PLoS Pathogens, 7, e1002068. 10.1371/journal.ppat.1002068
- Werner, O., Starick, E., Teifke, J., Klopfleisch, R., Prajitno, T.Y., Beer, M., Hoffmann, B. & Harder, T.C. (2007). Minute excretion of highly pathogenic avian influenza virus A/chicken/Indonesia/2003 (H5N1) from experimentally infected domestic pigeons (Columbia livia) and lack of transmission to sentinel chickens. Journal of General Virology, 88, 3089–3093. 10.1099/vir.0.83105-0