Abstract
For over three decades, there has been a continuing panzootic caused by a virulent variant avian paramyxovirus type 1 strain, the so-called pigeon paramyxovirus type 1. It is found primarily in racing pigeons, but it has also spread to wild birds and poultry. In this study, two pigeon paramyxovirus type 1 strains, SD12 and BJ13, obtained from diseased pigeons in China, were characterized. Phylogenetic analysis based on complete sequences allowed characterization of both strains as genotype VI, class II. Further phylogenetic analysis of a 374-nucleotide section of the fusion gene showed that SD12 fell into lineage VIbii-d and BJ13 into VIbii-f. The deduced amino acid sequence of the cleavage site of the fusion protein confirmed that both isolates contained the virulent motif 112K/RRQKR↓F117 at the cleavage site. Nevertheless, the values of intracerebral pathogenicity indices showed the SD12 isolate to be a velogenic strain and BJ13 isolate to be a mesogenic strain. The SD12 isolate was further investigated via clinical observation, RNA detection, histopathology and viral serology in experimentally infected 3-week-old chickens. It showed a mild pathological phenotype in chickens, with viral replication restricted to a few tissues. The molecular mechanism for the SD12 isolate to have a virulent motif but low levels of virulence for chickens requires further study.
Introduction
Newcastle disease (ND) is a highly contagious viral disease of chickens. The causative agent is Newcastle disease virus (NDV), also called avian paramyxovirus type 1 (APMV-1), which is taxonomically classified in the genus Avulavirus of family Paramyxoviridae (Mayo, Citation2002). Its genome is a non-segmented, single-stranded, negative-sense RNA molecule of approximately 15 kb, consisting of six transcriptional units encoding at least six proteins: the nucleocapsid protein (NP), the phosphoprotein (P), the matrix protein (M), the fusion protein (F), the haemagglutinin-neuraminidase (HN) and the polymerase protein (L) (Czeglédi et al., Citation2006). At least one additional, non-structural protein (V) and possibly a second non-structural protein (W) are generated by means of mRNA editing during P gene transcription (Steward et al., Citation1993; Peeters et al., Citation2004).
Based on the severity of the disease in chickens, NDV isolates are categorized into five pathotypes: viscerotropic velogenic, neurotropic velogenic, mesogenic, lentogenic and asymptomatic enteric (Alexander, Citation2011). Some studies showed that, in all studied NDV strains, virulence in chickens is correlated with cleavage sites of the F protein of the virus (Nagai & Klenk, Citation1977). Lentogenic strains have a monobasic amino acid motif at the F cleavage site, 112G-R/K-Q-G-R↓L117, and they are cleaved extracellularly by trypsin-like proteases found in the respiratory and intestinal tract. Mesogenic and velogenic strains have a multibasic amino acid motif, 112R/G/K-R-Q/K-K/R-R↓F117, which can be recognized by ubiquitous host proteases. This makes it possible to spread systemically and produce fatal infections (Glickman et al., Citation1988; Ogasawara et al., Citation1992).
In addition to chickens, NDV can also infect a wide variety of other avian species, such as Columbidae like pigeons and doves. The causative agent, which was antigenically and genetically distinguishable from other APMV-1 viruses, was termed pigeon paramyxovirus type 1 (PPMV-1). The PPMV-1 outbreak is currently considered endemic worldwide (Abolnik et al., Citation2008; Kim et al., Citation2008; Guo et al., Citation2013; Pchelkina et al., Citation2013; Aldous et al., Citation2014). Clinical signs in pigeons consist mainly of diarrhoea and a series of nervous disorders (Cattoli et al., Citation2011). However, in chickens, some PPMV-1 strains show reduced virulence or even a complete lack of virulence. Occasionally, the only clinical signs of PPMV-1 infection in layer chickens are a drop in egg production, misshaped eggs and soft egg shells (Alexander & Parsons, Citation1984). All PPMV-1 strains examined to date were found contain a multibasic amino acid motif at the F cleavage site. Several studies have shown that they are potentially virulent (Collins et al., Citation1994; Kommers et al., Citation2003; Dortmans et al., Citation2011). This becomes apparent after serial passages in chickens, indicating that the viruses currently circulating among pigeon populations can cause ND outbreaks.
The PPMV-1 virus has spread to chickens in several countries, including Great Britain, where there were 22 outbreaks in unvaccinated chickens in 1984 as a result of using feed contaminated with faeces from infected feral pigeons (Alexander et al., Citation1984). Although there has never been an ND outbreak in poultry contributing to PPMV-1 variant isolates in China, several isolates have been obtained from pigeons and doves since 1998 (Yu et al., Citation2001; Liu et al., Citation2007; Qin et al., Citation2008; Guo et al., Citation2013). In these studies, most strains were characterized genotypically and pathotypically. NDV evolves continuously, and there is evidence of accelerated evolution. It is therefore important to clinicopathologically characterize new strains isolated during outbreaks, especially strains isolated from species other than chickens.
In this study, two isolates of PPMV-1 (D12 and BJ13) were characterized through sequencing, phylogenetic analysis and intracerebral pathogenicity indices (ICPIs). The SD12 isolate showed a higher ICPI value. It was selected for animal testing. Three-week-old chickens were inoculated via a combined intraocular and intranasal route. The resulting disease was characterized through clinical observation, RNA detection, histopathology and viral serology.
Materials and Methods
Animal and ethics statement
Ninety 3-week-old specific pathogen-free (SPF) chickens were used to compare the pathogenicity of two PPMV-1 isolates and a commercial vaccine strain. Chickens were kept in isolators at China Agricultural University throughout the experiment and the rearing facilities were approved by Beijing Administration Committee of Laboratory Animals under the leadership of the Beijing Association for Science and Technology, the approval ID is XYXK (Jing) 2013-0013. The study was carried out in strict accordance with the Guidance for the Care and Use of Laboratory Animals formulated by the Ministry of Science and Technology of the People’s Republic of China. The protocol was approved by the Animal Welfare and Ethical Censor Committee at China Agricultural University. At the end of the observation period, all survival chickens were anaesthetized by CO2 inhalation and euthanized by cervical dislocation. All surgery was performed under sodium pentobarbital anaesthesia, and all efforts were made to minimize suffering.
Viruses and the ICPI test
Two PPMV-1 isolates were isolated from diseased pigeon flocks in 2012 and 2013 from Shandong province and Beijing city of China, in which the infected pigeons manifest clinical signs of diarrhoea and a series of nervous disorders. The number of diseased pigeons was variable but generally 10–15% of birds in a loft. The strains were designated Pigeon/China/SD/2012 (abbreviated as SD12) and Pigeon/China/BJ/2013 (abbreviated as BJ13), respectively. The two viruses were purified three times using a plaque technique before being propagated in the allantoic cavities of 10-day-old SPF embryonated chicken eggs. Virus titres were expressed as 50% embryo infectious dose (EID50/ml) by the end-point method of Reed and Muench. The pathogenic potential for the two isolated viruses was evaluated using standard assay methods to determine ICPI in 1-day-old SPF chicks (Alexander et al., Citation1998). Vaccine strain LaSota (ICPI = 0.40) and virulent strain GM (A genotype VII NDV isolate, ICPI = 1.78) were used as reference strains for pathogenicity comparison in this study. All virus stocks were stored at –80°C in the Key Laboratory of Animal Epidemiology and Zoonosis, Ministry of Agriculture, China Agricultural University (Beijing, China) until use.
Primer design and RNA extraction
Based on the available NDV nucleotide sequences IT-227/82 (AJ880277), NDV05-029 (FJ766528) and PPMV-1/Maryland/1984 (FJ410147), 22 pairs of specific primers were designed to amplify the complete genome sequences of SD12 and BJ13. All primers used for this study are listed in supplementary Table 1. Viral genomic RNA was extracted from allantoic fluid using Trizol reagent (Invitrogen, San Diego, CA, USA) according to the manufacturer’s instructions.
Table 1. Experimental infection: chicken groups and viruses used.
Reverse transcription-polymerase chain reaction
Reverse transcription was performed at 37°C for 1 h in a 20-μL reaction volume using 3 μg total RNA, 1 μL random primers (500 μg/mL random hexadeoxynucleotides; Promega, Madison, WI, USA) and 0.5 μL M-MLV reverse transcriptase (200 U/μL; Promega). The PCRs were performed in a thermocycler (Biometra, Goettingen, Germany) with 100-ng cDNA as template in a 20-μL reaction volume containing 10 pmol of each primer and 1-U Taq DNA polymerase (Promega). Reactions were performed according to the following protocol: 95°C for 5 min, followed by 35 cycles of 95°C for 45 s, 53°C or 55°C for 45 s, 72°C for 2 min, and a final elongation step of 10 min at 72°C (Zhang et al., Citation2010). PCR products were examined by electrophoresis on a 1.5% (w/v) agarose gel and visualized after Goldview staining.
Cloning and sequencing of PCR products
PCR products of the expected length were purified with a Gel Extraction kit (OMEGA, Norcross, GA, USA), then cloned into the PMD18-T vector (TaKaRa, Tokyo, Japan) according to the manufacturer’s instructions and sequenced at Sunbio Biotech (Beijing, China). At least three clones of each segment were sequenced to control for Taq DNA polymerase misincorporation errors.
Phylogenetic analysis
Complete NDV genomic sequences were obtained from GenBank (Supplementary Table 2). These included current vaccine strains, typical prevailing isolates in China, and reference strains for each known NDV genotype. These NDV sequences and the complete coding sequences of the two NDV isolates were aligned and analysed using the ClustalW multiple alignment algorithm in the MegAlign program in the DNASTAR software suite (version 3.1; DNAstar, Madison, WI, USA). A phylogenetic tree was constructed using MEGA4.0 software (Molecular Evolutionary Genetics Analysis, version 4.0) using the neighbour-joining method (1000 replicates for bootstrap). The evolutionary distances were computed using the pairwise distance method and maximum composite likelihood model (Tamura et al., Citation2007).
Table 2. Details of the two PPMV-1 isolates investigated in this study.
Clinicopathological assessment in chickens
As shown in , ninety 3-week-old SPF white Leghorn chickens were divided into seven groups (groups A, B, C, D, E, F and G). Group G was inoculated with phosphate buffered saline (PBS) as a negative control. The other six groups were inoculated via a combined intraocular and intranasal route with the appropriate virus strain for dissection and observation separately. Each bird received 105.0 EID50 of viral inoculum based on titrations in embryonated eggs to confirm the dose. Chickens were housed in isolators and food and water were provided ad libitum. The experiment was terminated at 21 days post-infection (dpi).
Groups D, E, F and G were observed daily for clinical signs (e.g., dishevelled feathers, diarrhoea, lethargy, fever or paralysis), morbidity and mortality for 21 days. Serum samples were collected on 0 and 21 dpi. The haemagglutination-inhibition (HI) assay was performed using standard methods and using four haemagglutinating units of antigen (Alexander et al., Citation1998). Cloacal swabs were taken from the surviving chickens of groups A, B, C and G on 0, 1, 3, 5 and 7 dpi, and were placed in PBS for reverse transcription-polymerase chain reaction (RT-PCR) to assess viral shedding. Two chickens each from group A, B and C and one chicken from group G were killed on 1, 3, 5, 7, 10, 15 and 20 dpi. Gross pathologic changes were observed, and tissues (heart, liver, spleen, lung, kidney, proventriculus, duodenum, pancreas, caecal tonsil, bursa of Fabricius, trachea, oesophagus, harderian glands, brain and thymus) were collected for viral RNA detection and histological analysis.
Viral RNA detection of tissue samples
Tissue samples were used for RT-PCR to determine tissue distribution of the virus. Total RNA was obtained by using Trizol Reagent. RT-PCR was performed as described above using a pair of primers (forward: CCCCCAGGTATCATATCGC; reverse: CTTGTAGTGGCTCTCATCTG), which amplify and detect a 446-bp fragment of the F gene of NDV. PCR products were analysed on 1.5% agarose gels.
Histopathology
Tissues were collected and fixed by immersion in 10% neutral formalin at room temperature for 48 h. Tissue was then routinely processed, embedded in paraffin wax and cut into 5-μm sections. The sections were stained with haematoxylin and eosin and examined for lesions attributable to NDV infection using light microscopy.
Results
ICPI test assessment of the two isolates
The ICPI scores of SD12 and BJ13 were 1.64 and 1.20, respectively, in chickens, indicating that these are both virulent strains based upon the World Organisation for Animal Health (OIE) international standards (Alexander, Citation2008). The ICPI scores and other details are shown in .
Sequence characteristics of the whole genome and phylogenetic analysis
The RT-PCRs performed with all primers (Supplementary Table 1) resulted in amplification of the expected products. The amplified products were sequenced, annotated and assembled to obtain the entire nucleotide sequences of two isolates. The accession numbers for the sequences of the coding regions of SD12 and BJ13 are KJ808820 and KJ808819, respectively. The genome of these two strains was 15,192 nucleotides (nt) in length. The gene order of 3′-N-P-M-F-HN-L-5′ coding from six open reading frames was similar to that of other APMV-1 strains. The coding region sequences of these two strains also exhibited a very low divergence (0.6–9.0%) from the classical genotype VI strains compared with the genotype II vaccine strains such as LaSota used in China. The length of the 3′ leader and 5′ trailer were 55 and 114 nt, respectively, as reported for most NDV strains.
Unlike NDV LaSota, both two isolates have a 6-nt insertion (CCCCAA) in positions 1647–1648 nt of the NP gene. Proteolytic cleavage site motifs (residues 112–117) for the F0 protein were analysed in the two isolates. Both strains were shown to have a virulent motif (112KRQKR↓F117 or 112RRQKR↓F117) composed of multibasic amino acids at the F0 cleavage site. This motif is commonly found in PPMV-1 strains (Alexander, Citation2011; Guo et al., Citation2013). Analysis of predicted N-glycosylation sites of F protein in the two PPMV-1 isolates revealed that besides six conserved potential sites located at positions 85N-R-T87, 191N-N-T193, 366N-T-S368, 447N-I-S449, 471N-N-S473 and 541N-N-T543, there was a seventh one located at position 497N-T-S499 in the SD12 isolate. Analysis of predicted N-glycosylation sites of the HN protein showed that five potential sites located at positions 119N-N-S121, 341N-N-T343, 433N-K-T435, 481N-H-T483 and 508N-I-S510 were conserved in the BJ13 isolate, but only four sites in the SD12 isolate, which lacked the fifth one at position 508N-I-S510. Further work is needed to explore the potential influence of these changes of numbers of N-glycosylation sites on virulence of strain SD12.
The nucleotide and amino acid sequences of the two isolates were compared. Strain SD12 and BJ13 shared 90.7% nucleotide identity in the coding region sequences. The six genes, NP, P, M, F, HN and L, shared 91.5%, 89.5%, 90.2%, 91.3%, 89.5%, 92.6% nucleotide identity and 96.3%, 90.2%, 96.2%, 94.6%, 93.5, 97.1% amino acid identity, respectively. Nucleotide sequence data of 94 NDV reference strains were obtained from the GenBank database (Supplementary Table 2) and used for comparison. The SD12 isolate was highly similar to China PPMV-1 isolate ND/05/028 (99.4%, Accession number GQ338311) obtained from Jiangsu Province. Strain BJ13 showed the greatest nucleotide identities (98.0%) with Pi/CH/LLN/110713 (Accession number JX486552), another recent domestic PPMV-1 strain isolated from Liaoning Province. It was also closely related to Belgium strains 11-09620 (Accession number JX901124) isolated in 2011 (98.5% nucleotide sequence homology). The two isolates (SD12 and BJ13) had sequence homologies of 77.9% and 82.2%, respectively, at the nucleotide level with strain LaSota (Accession number AF077761), which is a common vaccine strain used in China.
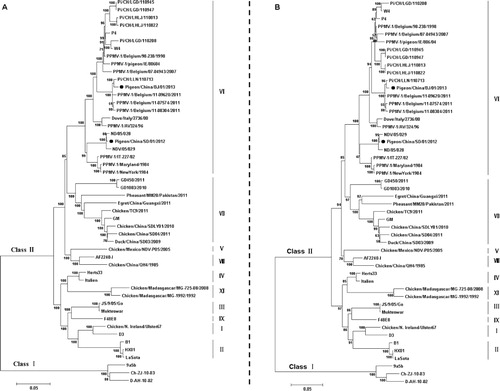
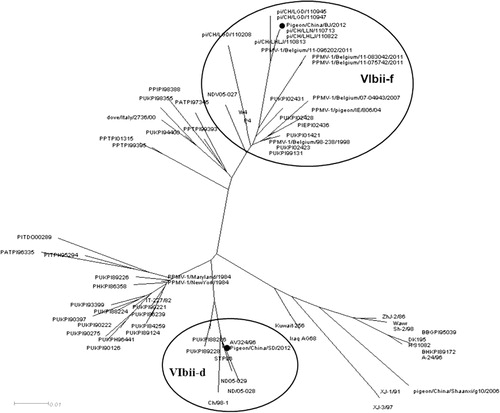
Phylogenetic analysis was conducted based on the complete genome sequences and sequences of the fusion gene of NDV. Both isolates were characterized as genotype VI (Ballagi-Pordány et al., Citation1996) as shown in . Further phylogenetic analysis based on a 374-nucleotide section of the fusion gene showed that the two PPMV-1 isolates examined in this study could be divided into two different subsets. The SD12 strain fell into clade VIbii-d, and the BJ13 isolate was identified as VIbii-f, as shown in . Both viruses were genetically distinct and phylogenetically distant from the vaccine strain LaSota.
Clinicopathological assessment in chickens
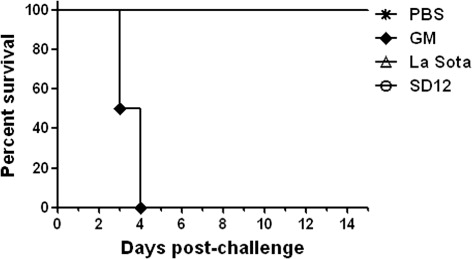
Chickens inoculated with SD12 showed no apparent clinical signs of infection. Chickens inoculated with the virulent strain GM showed classic clinical signs, such as dishevelled feathers, depression and paralysis. Starting on 3 dpi, these chickens began to die and all were dead on 5 dpi. Chickens inoculated with the LaSota vaccine strain did not show clinical signs and nor did the negative controls ().
At necropsy, chickens infected with SD12 showed slight gross lesions, such as a few off-white necrotic spots on spleen, and slight haemorrhaging of the caecal tonsil, thymus, proventriculus and duodenum. In contrast, chickens inoculated with virulent strain GM developed severe lesions in various organs, especially in digestive organs. Chickens in the vaccine strain control group showed minimal gross lesions, but the negative control group did not develop gross lesions.
Virus shedding
To determine virus shedding, cloacal swabs were taken on 0, 1, 3, 5 and 7 dpi. Cloacal swabs taken before inoculation were used to confirm the lack of PPMV-1 in chickens. The results are compiled in . Mock-inoculated chickens remained virus-negative throughout the experiment. Viral shedding was detectable in all three virus-inoculated groups but its period varied with the virulence of strains. Chickens infected with GM as virulent controls maintained a high rate of viral shedding until all chickens had died by 5 dpi. Like the vaccine controls, viral shedding in the SD12 group reached a peak on 1 dpi and had gradually declined by 5 dpi.
Table 3. Virus shedding in cloacal swabs as indicated by RT-PCR.
Serology
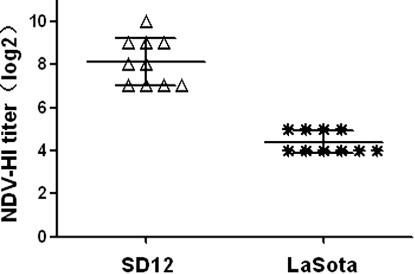
To evaluate the antibody responses of the infected chickens, serum samples were obtained from surviving chickens of SD12 group and LaSota group on 21 dpi, and HI tests were performed to detect specific NDV antibodies (). All sera were positive for NDV HI antibody after SD12 or LaSota inoculation, and average HI titers were 8.1 log2 for SD12 and 4.4 log2 for LaSota.
Viral RNA detection of tissue samples
To further examine the pathogenicity of SD12, the tissue tropism of the strain was investigated using RT-PCR. Fifteen kinds of tissue samples were collected. The results illustrated in showed that, in the SD12 group viral RNA was detected mainly in five kinds of tissue, including spleen, caecal tonsils, thymus, brain and kidney, but in the GM group it was detected in all tissue samples, indicating that the SD12 virus had a narrower tissue tropism than the GM virulent virus. The LaSota group was similar to the SD12 group in this respect.
Table 4. Viral distribution in tissues of chickens inoculated with SD12, GM and LaSota.
Histopathological analysis
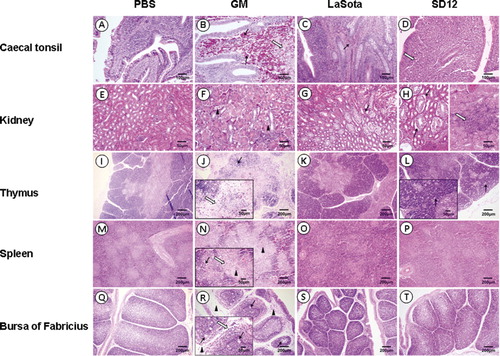
Chickens in SD12 and GM groups exhibited histopathological changes. The SD12 group showed less pronounced changes than the GM group. In the SD12 group, the spleen lesions () exhibited a starry sky change similar to that observed in the thymus lesions (). This was the result of the disruption and disappearance of a large number of lymphocytes. The amalgamation of collapsed cell and inflammatory exudates created the homogeneous and pink-stained appearance of white pulp in the spleen. The lesions in the kidney () were relatively slight. These lesions included cytomorphosis and detachment of basement membrane of renal tubular epithelial cells, renal tubular stenosis and local lymphocytic infiltration in the interstitium. In comparison, all tissues from the GM group presented more severe typical histological changes than tissues from other groups. Tissues from the LaSota group showed much milder histological changes, but none of the chickens in the negative control group had any apparent histological changes.
Discussion
Besides chickens, pigeons and doves, PPMV-1 viruses have also been isolated from budgerigars, cockatoos, falcons, kestrels, pheasants and swans (Biancifiori and Fioroni, Citation1983; Kaleta et al., Citation1985; Aldous et al., Citation2004; Duan et al., Citation2014). Clinical signs of pigeon infection include paralysis of the wings and legs, stiffening of the neck, excessive drinking and watery diarrhoea (Alexander & Parsons, Citation1984). In chickens, ICPI values for PPMV-1 are typical of mesogenic NDVs, but, in some cases, PPMV-1 isolates have shown increased virulence in chickens after several passages, causing them to pose a threat to poultry production. In this study, two NDV strains isolated from affected pigeons from northern China were characterized genotypically and pathotypically.
NDV strains consist of related but genetically, antigenically and phenotypically distinguishable viruses. Two classes of NDV are recognized (I and II). Class II viruses of NDV strains are categorized into genotypes I–XI with several sub-lineages within them. PPMV-1 isolates are characterized as VIb sub-lineage. In a recent study, considerable genetic variability has been demonstrated within the VIb sub-lineage and PPMV-1 viruses were divided into two distinguishable major genetic subgroups, VIbi and VIbii (Chong et al., Citation2013). Results suggested that the VIbi subgroup was diversified into three clades, termed “a”, “b” and “c”, while VIbii subgroup split into four clades, termed “d”, “e”, “f” and “g”. Previous work (Aldous et al., Citation2003) also showed that subgroup VIbii was the predominant strain in the latter period of this ongoing pigeon panzootic, and the number of subgroup VIbi isolates diminished starting in the late 1980s. In China, all PPMV-1 isolates fell into subgroup VIbii and were further divided into two clades (VIbii-d and VIbii-f). Several strains isolated at earlier times were characterized as clade VIbii-d, namely STP96 in 1996 (Qin et al., Citation2008), Ch/98-1 in 1998 (Yu et al., Citation2001), and NDV05-028 and NDV05-029 in 2005 (Liu et al., Citation2007). They clustered together with AV324/96 from Ireland, which was isolated in 1996. However, since then, this specific clade has been preserved and has evolved in pigeon flocks. Clade VIbii-f includes some recently identified strains, such as pi/CH/LGD/110947 and pi/CH/LLN/110713 (Guo et al., Citation2013), which were found in 2011. These Chinese isolates clustered together with strains found in Belgium in 2011. Our two isolates in this study share 90.7% nucleotide identity in the coding region sequences. In six genes, pairwise nucleotide sequence similarity between the two isolates ranged from 89.5% for P gene to 92.6% for L gene. The phylogenetic analysis based on the coding region sequences indicated that the two PPMV-1 isolates were classified as VIb sub-lineage. A phylogenetic tree based on a 374-bp fragment of an F gene placed the SD12 isolate in clade VIbii-d and BJ13 isolate in clade VIbii-f. This indicated that there were two distinct clusters that may have been introduced to China in two separate events. Viruses of clade VIbii-d may have been introduced into China by the importation of breeding or racing pigeons or wild birds some time during or after 1996. For a long time, viruses of this specific clade were preserved and evolved in pigeon flocks, leading to occasional outbreaks. Clade VIbii-f was probably introduced to China more recently and is currently circulating.
Most of the NDV strains isolated from pigeons do not result in significant disease in poultry and are considered mesogenic due to their ICPI values of 0.7–1.5 in chickens. Pathogenicity tests showed the BJ13 isolate to be a mesogenic strain similar to other isolates characterized in previous studies (Alexander & Parsons, Citation1984). The ICPI score was 1.64 for the SD12 isolate, indicating that it is virulent and notifiable based upon OIE international standards (Liu et al., Citation2003; Abolnik et al., Citation2008; Kim et al., Citation2008; Alexander, Citation2011; Guo et al., Citation2013; Pchelkina et al., Citation2013; Aldous et al., Citation2014). The pathogenicity of strain SD12 was assessed in chickens using clinical signs, gross lesions, tissue distribution, viral shedding and histological changes. There were no clinical signs or gross lesions in chickens inoculated with strain SD12, which is consistent with previous studies (Guo et al., Citation2014). Shedding virus via the cloacal route was observed from 1 to 5 dpi. Although tracheal swabs were not collected, viral RNA was detected in the trachea by tissue PCR. As shown in , viruses were detected in the trachea in one of two birds on 1, 5 and 7 dpi, indicating that viral shedding via the oral route may also start at 1 dpi, then lasting about one week, even though individual differences were found in chickens inoculated with SD12. SD12 showed less efficient viral replication in chickens than GM in all types of tissues. It is here suggested that the relatively low rate of viral replication and the relatively high antibody level in chickens may have contributed to the low pathogenicity. Dortmans et al. (Citation2010) reported that PPMV-1 virulence in chickens is directly associated with the activity of the viral replication. The current results confirmed the existence of differences in pathogenicity and efficiency of viral RNA replication, indicating different susceptibilities across different host species.
PPMV-1 presents a significant ongoing threat to domestic and wild bird populations. This study characterizes two recently isolated strains (SD12 and BJ13) genotypically and pathotypically. The information may facilitate the development of appropriate prevention and control measures. The PPMV-1 detected in pigeon populations in China merit continued investigation of PPMV-1 viruses that can spread from pigeons to chickens.
Supplemental data
Supplemental data for this article can be accessed here.
Supplemental data.pdf
Download PDF (237 KB)Additional information
Funding
References
- Abolnik, C., Gerdes, G.H., Kitching, J., Swanepoel, S., Romito, M. & Bisschop, S.P. (2008). Characterization of pigeon paramyxoviruses (Newcastle disease virus) isolated in South Africa from 2001 to 2006. The Onderstepoort Journal of Veterinary Research, 75, 147–152.
- Aldous, E.W., Fuller, C.M., Mynn, J.K. & Alexander, D.J. (2004). A molecular epidemiological investigation of isolates of the variant avian paramyxovirus type 1 virus (PPMV-1) responsible for the 1978 to present panzootic in pigeons. Avian Pathology, 33, 258–269. 10.1080/0307945042000195768
- Aldous, E.W., Fuller, C.M., Ridgeon, J.H., Irvine, R.M., Alexander, D.J. & Brown, I.H. (2014). The evolution of pigeon paramyxovirus type 1 (PPMV-1) in Great Britain: a molecular epidemiological study. Transboundary and Emerging Diseases, 61, 134–139. 10.1111/tbed.12006
- Aldous, E.W., Mynn, J.K., Banks, J. & Alexander, D.J. (2003). A molecular epidemiological study of avian paramyxovirus type 1 (Newcastle disease virus) isolates by phylogenetic analysis of a partial nucleotide sequence of the fusion protein gene. Avian Pathology, 32, 239–257. 10.1080/030794503100009783
- Alexander, D.J. (2008). Newcastle Disease. Chapter 2.3.14 World Organisation for Animal Health (OIE) Manual of Diagnostic Tests and Vaccines for Terrestrial Animals 6th edn (pp. 576–589). Paris: OIE.
- Alexander, D.J. (2011). Newcastle disease in the European Union 2000 to 2009. Avian Pathology, 40, 547–558. 10.1080/03079457.2011.618823
- Alexander, D.J. & Parsons, G. (1984). Avian paramyxovirus type 1 infections of racing pigeons: 2 pathogenicity experiments in pigeons and chickens. Veterinary Record, 114, 466–469. 10.1136/vr.114.19.466
- Alexander, D.J., Parsons, G. & Marshall, R. (1984). Infection of fowls with Newcastle disease virus by food contaminated with pigeon faeces. The Veterinary Record, 115, 601–602.
- Alexander, D.J., Purchase, H.G., Arp, L.H., Domermuth, C.H. & Pearson, J.E. (1998). A Laboratory Manual for the Isolation and Identification of Avian Pathogens. Newcastle Disease and Other Avian Paramyxovirus 4th edn (pp. 156–163). Kennett Square, PA: American Association of Avian Pathologists.
- Ballagi-Pordány, A., Wehmann, E., Herczeg, J., Belák, S. & Lomniczi, B. (1996). Identification and grouping of Newcastle disease virus strains by restriction site analysis of a region from the F gene. Archives of Virology, 141, 243–261. 10.1007/BF01718397
- Biancifiori, F. & Fioroni, A. (1983). An occurrence of Newcastle disease in pigeons: virological and serological studies on the isolates. Comparative Immunology, Microbiology and Infectious Diseases, 6, 247–252.
- Cattoli, G., Susta, L., Terregino, C. & Brown, C. (2011). Newcastle disease: a review of field recognition and current methods of laboratory detection. Journal of Veterinary Diagnostic Investigation, 23, 637–656. 10.1177/1040638711407887
- Chong, Y.L., Lam, T.T., Kim, O., Lu, H., Dunn, P. & Poss, M. (2013). Successful establishment and global dispersal of genotype VI avian paramyxovirus serotype 1 after cross species transmission. Infection, Genetics and Evolution, 17, 260–268. 10.1016/j.meegid.2013.04.025
- Collins, M.S., Strong, I. & Alexander, D.J. (1994). Evaluation of the molecular basis of pathogenicity of the variant Newcastle disease viruses termed “pigeon PMV-1 viruses”. The Journal of General Virology, 134, 403–411.
- Czeglédi, A., Ujvári, D., Somogyi, E., Wehmann, E., Werner, O. & Lomniczi, B. (2006). Third genome size category of avian paramyxovirus serotype 1 (Newcastle disease virus) and evolutionary implications. Virus Research, 120, 36–48. 10.1016/j.virusres.2005.11.009
- Dortmans, J.C., Rottier, P.J., Koch, G. & Peeters, B.P. (2010). The viral replication complex is associated with the virulence of Newcastle disease virus. Journal of Virology, 84, 10113–10120.
- Dortmans, J.C., Rottier, P.J., Koch, G. & Peeters, B.P. (2011). Passaging of a Newcastle disease virus pigeon variant in chickens results in selection of viruses with mutations in the polymerase complex enhancing virus replication and virulence. Journal of General Virology, 92, 336–345. 10.1099/vir.0.026344-0
- Duan, X., Zhang, P., Ma, J., Chen, S., Hao, H., Liu, H., Fu, X., Wu, P., Zhang, D., Zhang, W., Du, E. & Yang, Z. (2014). Characterization of genotype IX Newcastle disease virus strains isolated from wild birds in the northern Qinling Mountains, China. Virus Genes, 48, 48–55. 10.1007/s11262-013-0987-y
- Glickman, R.L., Syddall, R.J., Iorio, R.M., Sheehan, J.P. & Bratt, M.A. (1988). Quantitative basic residue requirements in the cleavage-activation site of the fusion glycoprotein as a determinant of virulence for Newcastle disease virus. Journal of Virology, 62, 354–356.
- Guo, H., Liu, X., Han, Z., Shao, Y., Chen, J., Zhao, S., Kong, X. & Liu, S. (2013). Phylogenetic analysis and comparison of eight strains of pigeon paramyxovirus type 1 (PPMV-1) isolated in China between 2010 and 2012. Archives of Virology, 158, 1121–1131.
- Guo, H., Liu, X., Xu, Y., Han, Z., Shao, Y., Kong, X. & Liu, S. (2014). A comparative study of pigeons and chickens experimentally infected with PPMV-1 to determine antigenic relationships between PPMV-1 and NDV strains. Veterinary Microbiology, 168, 88–97. 10.1016/j.vetmic.2013.11.002
- Kaleta, E.F., Alexander, D.J. & Russell, P.H. (1985). The first isolation of the avian PMV-1 virus responsible for the current panzootic in pigeons? Avian Pathology, 14, 553–557. 10.1080/03079458508436258
- Kim, L.M., King, D.J., Guzman, H., Tesh, R.B., Travassos da Rosa, A.P., Bueno, R. Jr., Dennett, J.A. & Afonso, C.L. (2008). Biological and phylogenetic characterization of pigeon paramyxovirus serotype 1 circulating in wild North American pigeons and doves. Journal of Clinical Microbiology, 46, 3303–3310. 10.1128/JCM.00644-08
- Kommers, G.D., King, D.J., Seal, B.S. & Brown, C.C. (2003). Virulence of six heterogeneous-origin Newcastle disease virus isolates before and after sequential passages in domestic chickens. Avian Pathology, 32, 81–93. 10.1080/0307945021000070750
- Liu, H., Wang, Z., Wu, Y., Zheng, D., Sun, C., Bi, D., Zuo, Y. & Xu, T. (2007). Molecular epidemiological analysis of Newcastle disease virus isolated in China in 2005. Journal of Virological Methods, 140, 206–211. 10.1016/j.jviromet.2006.10.012
- Liu, X.F., Wan, H.Q., Ni, X.X., Wu, Y.T. & Liu, W.B. (2003). Pathotypical and genotypical characterization of strains of Newcastle disease virus isolated from outbreaks in chicken and goose flocks in some regions of China during 1985–2001. Archives of Virology, 148, 1387–1403.
- Mayo, M.A. (2002). A summary of taxonomic changes recently approved by ICTV. Archives of Virology, 147, 1655–1656. 10.1007/s007050200039
- Nagai, Y. & Klenk, H.-D. (1977). Activation of precursors to both glycoproteins of Newcastle disease virus by proteolytic cleavage. Virology, 77, 125–134. 10.1016/0042-6822(77)90412-3
- Ogasawara, T., Gotoh, B., Suzuki, H., Asaka, J., Shimokata, K., Rott, R. & Nagai, Y. (1992). Expression of factor X and its significance for the determination of paramyxovirus tropism in the chick embryo. EMBO Journal, 11, 467–472.
- Pchelkina, I.P., Manin, T.B., Kolosov, S.N., Starov, S.K., Andriyasov, A.V., Chvala, I.A., Drygin, V.V., Yu, Q., Miller, P.J. & Suarez, D.L. (2013). Characteristics of pigeon paramyxovirus serotype-1 isolates (PPMV-1) from the Russian Federation from 2001 to 2009. Avian Diseases, 57, 2–7. 10.1637/10246-051112-Reg.1
- Peeters, B., Verbruggen, P., Nelissen, F. & de Leeuw, O. (2004). The P gene of Newcastle disease virus does not encode an accessory X protein. Journal of General Virology, 85, 2375–2378. 10.1099/vir.0.80160-0
- Qin, Z.M., Tan, L.T., Xu, H.Y., Ma, B.C., Wang, Y.L., Yuan, X.Y. & Liu, W.J. (2008). Pathotypical characterization and molecular epidemiology of Newcastle disease virus isolates from different hosts in China from 1996 to 2005. Journal of Clinical Microbiology, 46, 601–611. 10.1128/JCM.01356-07
- Steward, M., Vipond, I.B., Millar, N.S. & Emmerson, P.T. (1993). RNA editing in Newcastle disease virus. Journal of General Virology, 74, 2539–2547. 10.1099/0022-1317-74-12-2539
- Tamura, K., Dudley, J., Nei, M. & Kumar, S. (2007). MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Molecular Biology and Evolution, 24, 1596–1599. 10.1093/molbev/msm092
- Yu, L., Wang, Z., Jiang, Y., Chang, L. & Kwang, J. (2001). Characterization of newly emerging Newcastle disease virus isolates from the People’s Republic of China and Taiwan. Journal of Clinical Microbiology, 39, 3512–3519. 10.1128/JCM.39.10.3512-3519.2001
- Zhang, R., Wang, X., Su, J., Zhao, J. & Zhang, G. (2010). Isolation and analysis of two naturally-occurring multi-recombination Newcastle disease viruses in China. Virus Research, 151, 45–53. 10.1016/j.virusres.2010.03.015