ABSTRACT
During routine diagnosis in 2012, 69 samples of diseased turkey breeding and fattening flocks in Germany were examined for infection with aviadenoviruses by virus isolation using primary chicken embryo liver cells. In total, 21 aviadenovirus isolates, identified by a group-specific indirect immunofluorescence test, were obtained from 19 flocks. In almost all cases, molecular typing of these isolates based on partial hexon gene sequences revealed the presence of different types of turkey aviadenoviruses (TAdVs), including species Turkey aviadenovirus B (TAdV-B) with at least two different genotypes, as well as the species Turkey aviadenovirus C (TAdV-C) and Turkey aviadenovirus D (TAdV-D). Further analysis of DNA-dependent DNA polymerase gene sequences confirmed the classification of selected TAdV-C and TAdV-D isolates. Based on the results obtained for both genes, we suggest that TAdV-2, in addition to TAdV-4, belongs to the species TAdV-C. In contrast, amplification of the DNA polymerase gene fragment of nearly all investigated TAdV-B isolates failed due to unknown reasons. The results of sequence and phylogenetic analysis support the previously proposed classification of TAdVs into three different species and demonstrated how widely spread these viruses are in German turkey flocks. Analysis of case histories revealed a wide range of clinical and pathological changes; however an apparent link between types and disease conditions was not identified.
Introduction
Avian adenoviruses exhibit a worldwide distribution in commercial poultry and in wild birds (Smyth & McNulty, Citation2008). Within the family Adenoviridae, two of the five genera, namely Aviadenovirus and Siadenovirus, cause infections in turkeys.
The genus Aviadenovirus comprises, amongst others, the fowl aviadenovirus (FAdV) and the turkey aviadenovirus (TAdV) species (Harrach et al., Citation2011; Harrach & Kajan, Citation2011). Species classification is based on phylogeny, genome organization and restriction fragment length polymorphism, whereas serotyping is performed via cross-neutralization tests (Hess, Citation2000; Harrach et al., Citation2011). Therefore, FAdVs are grouped into the five species Fowl aviadenovirus A to Fowl aviadenovirus E (FAdV-A to FAdV-E) and into 12 serotypes (FAdV-1 to FAdV-8a and FAdV-8b to FAdV-11) (Harrach et al., Citation2011). Furthermore, FAdVs can be grouped into at least 12 genotypes based on partial hexon gene sequences (Marek et al., Citation2010). To date, three TAdV species are known to exist within the genus Aviadenovirus. Turkey aviadenovirus B (TAdV-B), type TAdV-1, was isolated in Hungary from birds showing respiratory signs (Kajan et al., Citation2010). Very recently, two new species were described, Turkey aviadenovirus C (TAdV-C) with type TAdV-4, isolated in Northern Ireland and originally designated as TAdV-1, and Turkey aviadenovirus D (TAdV-D) with type TAdV-5 which comprises two strains isolated in the UK and Hungary (Marek et al., Citation2014a). In addition, a further type, TAdV-2, which was isolated in Northern Ireland, has also been suggested to belong to the genus Aviadenovirus (Kajan et al., Citation2010).
The genus Siadenovirus comprises various species infecting not only birds but also reptiles. The species Turkey siadenovirus A (TAdV-A), type TAdV-3, is associated with specific diseases of several avian hosts such as haemorrhagic enteritis in turkeys (Harrach et al., Citation2011). In contrast, aviadenoviruses are often detected in apparently healthy birds. However, some strains are more virulent and may cause quail bronchitis, inclusion body hepatitis, hydropericardium hepatitis syndrome and gizzard erosions in chickens (Gjevre et al., Citation2013; Hess, Citation2013). The pathogenicity of aviadenoviruses in turkeys remains unclear to date. These viruses have been isolated in the context of respiratory disease and poult enteritis complex as well as in association with inclusion body hepatitis in turkey poults and also may be responsible for lower hatchability rates (Guy et al., Citation1988; Shivaprasad et al., Citation2001; Hess, Citation2013; Moura-Alvarez et al., Citation2013). Despite their suggested clinical association, the overall prevalence of aviadenoviruses in turkey flocks is not well documented. Older case reports described adenovirus detection in turkeys in North Ireland and the USA (Scott & McFerran, Citation1972; Blalock et al., Citation1975; Simmons et al., Citation1976; Dillman & Simmons, Citation1977; Guy et al., Citation1988; Crespo et al., Citation1998). In Brazil, the occurrence of aviadenoviruses in turkeys was demonstrated in connection with poult enteritis complex (Moura-Alvarez et al., Citation2013). In addition, infections with aviadenoviruses in turkeys were also reported in the UK and Hungary (Kajan et al., Citation2010; Marek et al., Citation2014a). Unfortunately, the species or serotype was not determined in most of these publications. The present paper reports the results of the aviadenovirus isolation and identification of routine diagnostic samples originating from German breeding and fattening turkey flocks. Further characterization was performed by sequence and phylogenetic analysis of partial hexon gene and partial DNA-dependent DNA polymerase gene.
Materials and methods
Samples
In 2012, mixed tissue samples from 69 German turkey flocks were submitted for routine diagnostic virus isolation. The samples originated from turkey breeder and fattening flocks predominantly presenting with enteritis followed by hepatitis, splenitis and tenosynovitis. In one flock, increased mortality and hydropericardium syndrome were observed. In 24 flocks, no case history was reported.
Virus isolation and identification
Tissue samples were homogenized either individually or as a pool in phosphate buffered saline (PBS) as 20% solution. After three freeze–thaw cycles, homogenates were centrifuged for 15 min at 3000 x g and supernatants were filtered through 0.45 and 0.2 µm pore size filters. Subsequently, confluent chicken embryo liver (CEL) cell monolayers prepared from 11-day-old specific pathogen free chicken embryos were inoculated with the supernatants. Cell cultures were incubated at 37°C and 5% CO2. Monolayers were monitored daily for cytopathogenic effects (CPE). If no CPE was present after four to five days, up to three additional passages were performed.
For virus identification, a group-specific indirect immunofluorescence test was carried out (Adair et al., Citation1980). Primary CEL cells, grown on cover slips in cell culture dishes, were inoculated with the last CPE-inducing passage on CEL cells. After a 24-h incubation period, coverslips were fixed with acetone for 8 min and stained with 100 µl anti-FAdV-1 rabbit serum (Intervet GmbH, Unterschleißheim, Germany). After incubation for 30 min at room temperature, infected cells were washed several times using PBS and incubated for further 30 min at room temperature with 100 µl goat anti-rabbit serum conjugated to fluorescein isothiocyanate (Bio-Rad AbD Serotec GmbH, Puchheim, Germany). The coverslips were washed again in PBS and analysed by fluorescence microscopy.
DNA extraction
Automated DNA extraction from infected cells was performed using the QIAamp DNA Mini QIAcube Kit® (Qiagen GmbH, Hilden, Germany) according to the manufacturer’s instructions.
Polymerase chain reaction and sequencing
Molecular typing was carried out by amplification of all obtained isolates using a conventional polymerase chain reaction (PCR) based on the L1 region of the hexon gene with an approximate product size of 897 bp (Meulemans et al., Citation2001). Amplification was performed using PuRe Taq Ready-To-Go PCR Beads® (GE Healthcare, Munich, Germany). Each reaction volume was made up of 25 µl containing 0.4 µM of forward (hexon A 5ʹ-CAARTTCAGRCAGACGGT-3ʹ) and reverse (hexon B 5ʹ-TAGTGATGMCGSGACATCAT-3ʹ) primers and 10 µl of extracted DNA. Initial denaturation lasted for 2 min at 94°C and was followed by 40 cycles of denaturation at 94°C for 1 min, annealing at 62°C for 1 min and extension at 72°C for 1.5 min. The final extension step lasted for 2 min at 72°C. Additionally, for selected isolates representing TAdV-B, TAdV-C and TAdV-D, a 1236 bp fragment within the DNA polymerase gene was analysed. Primer selection was performed by primer-BLAST (National Center for Biotechnology Information, Bethesda, MD, USA) on the basis of pairwise alignment of DNA polymerase sequences of TAdV-4 (KF477312) and TAdV-5 (KF477313) (Ye et al., Citation2012). Amplification was carried out using PuRe Taq Ready-To-Go PCR Beads® in a total volume of 25 µl containing 0.4 µM of forward (DNAPol-f 5ʹ-CCTCGTGACGCTGCCAA-3ʹ) and reverse (DNAPol-r 5ʹ-AACGAAGCGCTATACGACGA-3ʹ) primers and 5 µl of extracted DNA with the following cyclic conditions: initial denaturation at 94°C for 2 min, 30 cycles consisting of denaturation at 94°C for 15 s, annealing at 50°C for 30 s and elongation at 72°C for 1.5 min followed by final extension at 72°C for 7 min. Amplification products were analysed by electrophoresis in 1% agarose gel, stained with ethidium bromide and visualized by UV illumination. After electrophoresis PCR products were purified using the MinElute PCR Purification Kit® according to the manufacturer’s recommendations (Qiagen GmbH, Hilden, Germany) and sequenced directly in both directions using the hexon- and DNA polymerase gene-specific primers by a commercial sequencing service (LGC Genomics GmbH, Berlin, Germany). A subset of samples was sequenced in a microtitre plate format. In these cases, clean-up treatment of PCR products was performed by the sequencing service. Nucleotide sequences were submitted to GenBank.
Sequence and phylogenetic analysis
The nucleotide sequences of the approximately 897-bp region (amplified by primers hexon A and hexon B) and of the 1236-bp region (amplified by primers DNAPol-f and DNAPol-r) were analysed using Bioedit (version 7.0.0) (Ibis Biosciences, Carlsbad, CA, USA). For sequence analyses, primer binding sites were excluded and resulting sequences were translated to amino acid sequences. These sequences were trimmed to identical length and their alignment was executed via the MUSCLE algorithm. Pairwise identities were analysed using MEGA software, version 6.06 (Center for Evolutionary Medicine and Informatics, Tempe, AZ, USA), based on distance matrices of the obtained amino acid sequences of 196 aa (hexon) and 430 aa (DNA-dependent DNA polymerase) length (Tamura et al., Citation2013). For further typing, selected isolates were subjected to multiple sequence alignments and pairwise identity analysis based on distance matrices of nucleotide sequences of the partial hexon gene nucleotide sequences (544 nt) using the same methods as described above. Phylogenetic calculations of the amino acid sequences were performed using the maximum likelihood method online by Topali, version 2.5 (Biomathematics and Statistics Scotland, Edinburgh, UK). The resulting phylogenetic trees were visualized by the MEGA program.
Accession numbers
GenBank accession numbers were assigned to the nucleotide sequences of partial hexon gene and partial DNA-dependent DNA polymerase gene sequences as follows: GB 1005/12: KP828387, KR014810; GB 1006/12: KP828386; GB 1136/12: KP828385; GB 1592/12: KP828384, KR014809; GB 1643/12: KP828383; GB 1729/12: KP828367; GB 1882/12 tendons: KP323381; GB 1882/12 intestine: KP828380; GB 2051/12 pool: KP828379, KR014812; GB 2051/12 intestine: KP828379, KR014813; GB 2071/12: 828378; GB 2082/12: KP828377; GB 2081/12: KP828376; GB 2085/12: KP828375, KR014811; GB 2100/12: KP828374; GB 2150/12: KP828373, KR014808; GB 2203/12: KP828372, KR014807; GB 2331/12: KP828371; GB 2853/12: KP828370; GB 3158/12: KP828369, KR014806; GB 3293/12: KP828368.
Results
Isolation and identification
In total, 21 aviadenovirus isolates from 19 flocks were recovered, all showing a CPE after the first to fourth passage with small rounded cell degeneration and detachment from the surface which is known to be typical for adenoviruses. Group-specific indirect immunofluorescence test identified all isolates as members of the genus Aviadenovirus. Isolation yielded best results using intestinal and caecal tonsil tissue. In two cases, aviadenoviruses were isolated from tendons. Furthermore, tissue samples from certain flocks all showed successful virus isolation, whilst other flocks rendered only one positive sample (see supplementary data).
Sequence and phylogenetic analysis on partial hexon gene
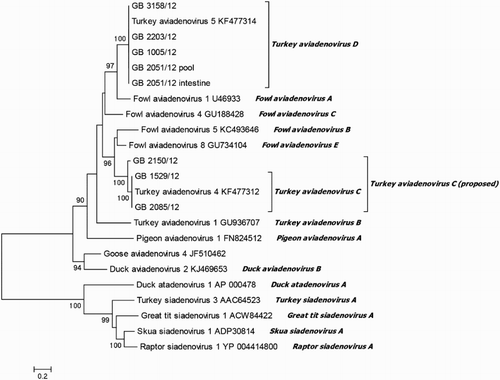
For further typing, a conventional PCR was performed to amplify a fragment within the L1 region of the hexon gene. For all 21 isolates, a fragment of the expected size of about 897 bp was obtained. Subsequently, PCR products were subjected for sequence analysis and deduced amino acid sequences were used for phylogenetic analysis in comparison to published avian adenovirus sequences. Seven of the 21 isolates were found to cluster with TAdV-B (), showing 90.2–99.3% amino acid sequence identities to TAdV-1 and 88.8–100% amongst themselves (). These findings were confirmed by nucleotide sequence analysis which revealed comparable results (). Nine of the obtained isolates clustered with TAdV-D, displaying 100% amino acid sequence identity to TAdV-5 and also amongst themselves. Three other isolates clustered with TAdV-C, two of them showed 98.6% identity to TAdV-4 with 100% identity amongst themselves, whereas the third isolate displayed 100% identity to TAdV-2. The comparison of the nucleotide sequences of these isolates showed 77.0–100% identity to each other. The two remaining isolates were found to cluster with FAdV-B and FAdV-E, showing only 59.4–72.7% amino acid sequence identity to TAdV species, respectively, and 60.7% identity amongst each other. However, these isolates displayed a relatively high sequence identity of 90.9% and 98.6% to FAdV-5 and FAdV-8b, respectively (data not shown). In 17 cases, one isolate per flock was detected, whereas in two flocks, “GB 1882/12” and “GB 2051/12”, two different species were identified. In both of these cases, TAdV-B was obtained from intestine, whereas TAdV-D was isolated from pools composed of various organ samples.
Figure 1. Phylogenetic tree based on amino acid sequence of partial hexon gene (196 aa). Online available sequences were compared to the field isolates. Branch lengths are given in number of substitutions per site (see the scale).
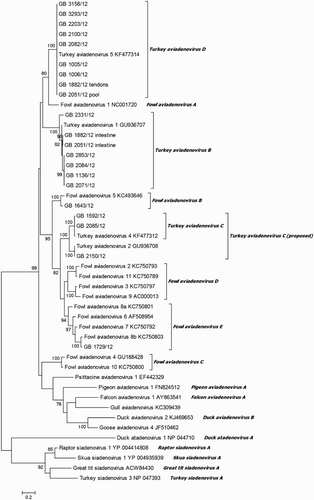
Table 1. Pairwise identity of partial hexon gene on nucleotide and amino acid level of TAdV-B isolates (544 nt and 196 aa).
Sequence and phylogenetic analysis on partial DNA-dependent DNA polymerase gene
Amino acid sequences of partial DNA-dependent DNA polymerase gene were analysed in comparison with published sequences for confirmation of previous results. Eleven isolates, representing the species TAdV-B, TAdV-C and TAdV-D, were tested. For seven isolates, classified by hexon gene analysis as TAdV-C and TAdV-D, respectively, a fragment of the expected size of about 1236 bp was obtained. Phylogenetic analysis confirmed their classification as TAdV-C and TAdV-D, respectively (). In addition, analysis of amino acid sequence identity analysis verified these results by 100% amino acid sequence identity of TAdV-D-classified isolates to TAdV-5. TAdV-C isolates showed 100% as well as 93.8% identity to TAdV-4. Unfortunately, a published DNA-dependent DNA polymerase gene reference sequence for TAdV-2 was not available. Amplification of partial DNA-dependent DNA polymerase gene of isolates representative for TAdV-B was only successful for one isolate. The isolate “GB 2051/12 intestine” showed 67.8% identity to TAdV-1 and 100% identity to TAdV-5 on partial DNA-dependent DNA polymerase gene. This isolate was additionally sequenced twice with a similar result.
Isolated types and case histories
Isolation of TAdV-B and TAdV-C (type 2 and 4) was successful from turkey flocks clinically presenting with enteritis and poor growth, respectively. TAdV-D (type 5), FAdV-B and FAdV-E were observed in flocks displaying hepatitis, splenitis, enteritis and polyserositis. Moreover, TAdV-D was isolated from one flock with turkeys showing hydropericardium syndrome and increased mortality.
Discussion
Aviadenoviruses are known to be globally distributed in chicken flocks; however detailed reports concerning their prevalence in turkeys are rare (Hess, Citation2013). With this background the present paper reveals how widely aviadenoviruses are distributed in German turkey flocks. In general, the use of cell cultures derived from the homologous species is described as preferable for aviadenovirus isolation (Hess, Citation2013). According to this, it was reported before that some turkey adenoviruses grow solely in primary cells of turkey origin (Scott & McFerran, Citation1972). Nevertheless, adenoviruses have also been isolated successfully from various avian species including turkeys using chicken cells (Niczyporuk, Citation2016). In the present investigation, virus isolation was successful in 19 of 69 turkey flocks examined using the latter culturing method. All isolates were recovered on CEL cells displaying the characteristically rounded cell degeneration after the first to fourth passage. Indirect immunofluorescence analysis confirmed aviadenovirus classification. Unfortunately, for some of the reported adenovirus detections in turkeys, the investigation of the causative serotype or species is lacking (Simmons et al., Citation1976; Sutjipto et al., Citation1977; Saif et al., Citation1985; Crespo et al., Citation1998). In some cases, FAdV-1 (FAdV-A) as well as TAdV-4 (TAdV-C), originally designated as TAdV-1 and TAdV-2, were identified (Scott & McFerran, Citation1972; Blalock et al., Citation1975; Cho, Citation1976; Guy & Barnes, Citation1997). In addition, recent publications described TAdV-1 (TAdV-B) and TAdV-5 (TAdV-D) as newly identified types detected in turkeys (Kajan et al., Citation2010; Marek et al., Citation2014a). However, data concerning their prevalence and relevance in Europe are lacking. Hence, this study could demonstrate the presence of all three aviadenoviral TAdV species in German turkey flocks by partial hexon gene analysis, the detection of which was also possible in the following years (Dörte Lüschow, personal communication, November 9, 2015). FAdV species were detected only in two cases. Moreover, the majority of viruses isolated were identified as TAdV-D (type TAdV-5). These results do not only show how widely these species are spread within German turkey flocks, but also underline their previously suggested relevance in Europe (Marek et al., Citation2014a). In contrast to TAdV-D isolates, hexon gene analysis of TAdV-B isolates revealed a greater heterogeneity. For FAdV, the existence of at least 12 genotypes within FAdV-A to -E was reported. Genotype grouping was established via a threshold value of 95% for nucleotide sequence identity within a hexon gene fragment (Marek et al., Citation2010). Based on this criterion, classification of TAdV-B isolates into at least two different genotypes was possible.
Species demarcation criteria include phylogenetic distance based on amino acid sequence of the DNA polymerase gene (Harrach et al., Citation2011). In order to confirm the results of the hexon gene analysis, a larger fragment of the DNA-dependent DNA polymerase gene was further investigated for selected isolates. Outcomes of both analyses were almost equivalent for TAdV-C and TAdV-D isolates. One isolate, typed by hexon gene analysis as TAdV-2, showed 94.0% amino acid sequence identity to TAdV-4 within the DNA polymerase gene. To our knowledge, this is the first published detection of TAdV-2 since the 1980s (Guy et al., Citation1988; Guy & Barnes, Citation1997). Moreover, based on phylogenetic analysis of both investigated genes and supported by the same host range and pathogenicity (Harrach et al., Citation2011), we suggest that TAdV-2 belongs to TAdV-C species, representing a second genotype besides TAdV-4. Unfortunately, amplification of the DNA polymerase gene fragment of nearly all TAdV-B isolates failed. In addition, the use of further primers located in the same region and developed on the basis of the published TAdV-1 sequence was also not successful (data not shown). An additional analysis of the 52k gene region may be an option for further characterization (Gunes et al., Citation2013). An amplicon was detected only for one isolate (GB 2051/12 intestine). However, this isolate displayed a high sequence identity to TAdV-1 within the hexon gene and to TAdV-5 within the DNA polymerase gene. Furthermore, in this case one other isolate, identified as TAdV-D (type TAdV-5), was obtained from different specimens within the same flock. A dual infection with TAdV-B and TAdV-D could be a possible explanation for this result.
The present paper demonstrates the occurrence of all aviadenoviral TAdV species within diseased turkey flocks, despite no apparent link between case history and type of isolate. In contrast to many previous publications, respiratory disease did not appear to be present in these cases (Blalock et al., Citation1975; Sutjipto et al., Citation1977; Crespo et al., Citation1998; Kajan et al., Citation2010). However, the presence of aviadenoviruses in turkeys which suffered from enteritis or hepatitis was reported before (Cho, Citation1976; Guy & Barnes, Citation1997; Prusas & Hafez, Citation2002; Woolcock & Shivaprasad, Citation2008; Moura-Alvarez et al., Citation2013). Since only diseased turkey flocks were sampled, the pathogenicity of aviadenoviruses in turkeys remains unclear.
In conclusion, our results obtained by investigation of a large number of field isolates indicate a high prevalence of turkey adenoviruses in German turkey flocks and support the classification of turkey adenoviruses into three different species within the genus Aviadenovirus (Marek et al., Citation2014a).
Supplementary_Data.docx
Download MS Word (18.5 KB)Acknowledgements
We gratefully acknowledge all of our colleagues for providing us with tissue samples. The exceptional technical assistance of Regina Schurich is greatly appreciated.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Adair, B.M., McFerran, J.B. & Calvert, V.M. (1980). Development of a microtitre fluorescent antibody test for serological detection of adenovirus infection in birds. Avian Pathology, 9, 291–300. doi: 10.1080/03079458008418414
- Blalock, H.G., Simmons, D.G. & Muse, K.E. (1975). Adenovirus respiratory infection in turkey poults. Avian Diseases, 19, 707–716. doi: 10.2307/1589184
- Cho, B.R. (1976). An adenovirus from a turkey pathogenic to both chicks and turkey poults. Avian Diseases, 20, 714–723. doi: 10.2307/1589451
- Crespo, R., Shivaprasad, H.L., Droual, R., Chin, R.P., Woolcock, P.R. & Carpenter, T.E. (1998). Inclusion body tracheitis associated with avian adenovirus in turkeys. Avian Diseases, 42, 589–596. doi: 10.2307/1592687
- Dillman, R.C. & Simmons, D.G. (1977). Histopathology of a rhinotracheitis of turkey poults associated with adenoviruses. Avian Diseases, 21, 481–491. doi: 10.2307/1589406
- Gjevre, A.-G., Kaldhusdal, M. & Eriksen, G.S. (2013). Gizzard erosion and ulceration syndrome in chickens and turkeys: a review of causal or predisposing factors. Avian Pathology, 42, 297–303. doi: 10.1080/03079457.2013.817665
- Gunes, A., Marek, A. & Hess, M. (2013). Species determination of fowl adenoviruses based on the 52K gene region. Avian Diseases, 57, 290–294. doi: 10.1637/10323-081012-ResNote.1
- Guy, J.S. & Barnes, H.J. (1997). Characterization of an avian adenovirus associated with inclusion body hepatitis in day-old turkeys. Avian Diseases, 41, 726–731. doi: 10.2307/1592167
- Guy, J.S., Schaeffer, J.L. & Barnes, H.J. (1988). Inclusion-body hepatitis in day-old turkeys. Avian Diseases, 32, 587–590. doi: 10.2307/1590936
- Harrach, B., Benko, M., Both, G.W., Brown, M., Davison, A.J., Echavarria, M., Hess, M., Jones, M.S., Kajon, A., Lehmkuhl, H.D., Mautner, V., Mittal, S.K. & Wadell, G. (2011). Family Adenoviridae. In A.M.Q. King, E. Lefkowitz, M.J. Adams & E.B. Carstens (Eds.), Virus Taxonomy: 9th Report of the International Committee on Taxonomy of Viruses (pp. 125–141). New York: Elsevier.
- Harrach, B. & Kajan, G.L. (2011). Aviadenovirus. Adenoviridae. In C.A. Tidona & G. Darai (Eds.), The Springer Index of Viruses (2nd ed., Vols 4, pp. 13–28). New York: Springer-Verlag.
- Hess, M. (2000). Detection and differentiation of avian adenoviruses: a review. Avian Pathology, 29, 195–206. doi: 10.1080/03079450050045440
- Hess, M. (2013). Aviadenovirus infections. In D.E. Swayne, J.R. Glisson, L.R. McDougald, L.K. Nolan, D.L. Suare & V.L. Nair (Eds.), Diseases of Poultry (13th ed., pp. 290–300). Ames: Wiley-Blackwell.
- Kajan, G.L., Stefancsik, R., Ursu, K., Palya, V. & Benko, M. (2010). The first complete genome sequence of a non-chicken aviadenovirus, proposed to be turkey adenovirus 1. Virus Research, 153, 226–233. doi: 10.1016/j.virusres.2010.08.006
- Marek, A., Ballmann, M.Z., Kosiol, C., Harrach, B., Schlotterer, C. & Hess, M. (2014a). Whole-genome sequences of two turkey adenovirus types reveal the existence of two unknown lineages that merit the establishment of novel species within the genus aviadenovirus. Journal of General Virology, 95, 156–170. doi: 10.1099/vir.0.057711-0
- Marek, A., Gunes, A., Schulz, E. & Hess, M. (2010). Classification of fowl adenoviruses by use of phylogenetic analysis and high-resolution melting-curve analysis of the hexon L1 gene region. Journal of Virological Methods, 170, 147–154. doi: 10.1016/j.jviromet.2010.09.019
- Meulemans, G., Boschmans, M., Berg, T.P.v.d. & Decaesstecker, M. (2001). Polymerase chain reaction combined with restriction enzyme analysis for detection and differentiation of fowl adenoviruses. Avian Pathology, 30, 655–660. doi: 10.1080/03079450120092143
- Moura-Alvarez, J., Chacon, J.V., Scanavini, L.S., Nunez, L.F., Astolfi-Ferreira, C.S., Jones, R.C. & Piantino Ferreira, A.J. (2013). Enteric viruses in Brazilian turkey flocks: single and multiple virus infection frequency according to age and clinical signs of intestinal disease. Poultry Science, 92, 945–955. doi: 10.3382/ps.2012-02849
- Niczyporuk, J.S. (2016). Phylogenetic and geographic analysis of fowl adenovirus field strains isolated from poultry in Poland. Archives of Virology, 161, 33–42. doi: 10.1007/s00705-015-2635-4
- Prusas, C. & Hafez, H.M. (2002). Mixed infection of meat turkeys with Turkey Adenovirus and Reovirus. Proceedings of the 4th International Symposium on Turkey Diseases, Berlin.
- Saif, L.J., Saif, Y.M. & Theil, K.W. (1985). Enteric viruses in diarrheic turkey poults. Avian Diseases, 29, 798–811. doi: 10.2307/1590671
- Scott, M. & McFerran, J.B. (1972). Isolation of adenoviruses from turkeys. Avian Diseases, 16, 413–420. doi: 10.2307/1588807
- Shivaprasad, H.L., Woolcock, P.R. & McFarland, M.D. (2001). Group I avian adenovirus and avian adeno-associated virus in turkey poults with inclusion body hepatitis. Avian Pathology, 30, 661–666. doi: 10.1080/03079450120092152
- Simmons, D.G., Miller, S.E., Gray, J.G., Blalock, H.G. & Colwell, W.M. (1976). Isolation and identification of a turkey respiratory adenovirus. Avian Diseases, 20, 65–74. doi: 10.2307/1589474
- Smyth, J.A. & McNulty, M.S. (2008). Adenoviridae. In M. Pattison, P.F. McMullin, J.M. Bradbury & D.J. Alexander (Eds.), Poultry Diseases (6th ed., pp. 367–381). Edinburgh: Elsevier.
- Sutjipto, S., Miller, S.E., Simmons, D.G. & Dillman, R.C. (1977). Physicochemical characterization and pathogenicity studies of two turkey adenovirus isolants. Avian Diseases, 21, 549–556. doi: 10.2307/1589413
- Tamura, K., Stecher, G., Peterson, D., Filipski, A. & Kumar, S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Molecular Biology and Evolution, 30, 2725–2729. doi: 10.1093/molbev/mst197
- Woolcock, P.R. & Shivaprasad, H.L. (2008). Electron microscopic identification of viruses associated with poult enteritis in turkeys grown in California 1993–2003. Avian Diseases, 52, 209–213. doi: 10.1637/8106-090607-Reg.1
- Ye, J., Coulouris, G., Zaretskaya, I., Cutcutache, I., Rozen, S. & Madden, T.L. (2012). Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinformatics, 13, 134–144. doi: 10.1186/1471-2105-13-134