ABSTRACT
In recent years, avian influenza virus (AIV) and Newcastle disease virus (NDV) have caused large-scale outbreaks in many countries, including Egypt. The culling and vaccination strategies have failed to control both viruses in Egypt. In this study, we investigated the outbreaks of nervous manifestations and deaths in pigeons between 2013 and 2015. The H5N1 subtype of the highly pathogenic avian influenza virus and pigeon paramyxovirus-1, an antigenic variant of NDV, were found to be the cause; AIV and pigeon paramyxovirus-1 were isolated from 61.3% (19/31) and 67.8% (21/31) of tested pigeons, respectively. Co-infection with both viruses was detected in 51.6% of pigeons (16/31). The AIV sequences showed PQGEKRRKKR/GLF motif at the haemagglutinin gene cleavage site, which is typical of the highly pathogenic H5N1 subtype. The phylogenetic tree showed that the highly pathogenic avian influenza belonged to clade 2.2.1.2. The NDV sequences carried one of the three motifs, 112GKQGRL117, 112KRQKRF117 or 112RRQKRF117, at the fusion protein cleavage site and were classified as genotypes I, VI and II in NDV-class II, respectively. This indicated that different genotypes of NDV can circulate simultaneously among pigeons. Further analysis revealed the clustering of some sequences in sub-genotypes Ia and VIb.2. To the best of our knowledge, these sub-genotypes have not been previously reported from pigeons in Egypt. Our results should serve as a base for future studies on both viruses in Egypt.
KEYWORDS:
Introduction
Highly pathogenic avian influenza virus (HPAIV) and virulent Newcastle disease virus (NDV) cause important diseases in more than 250 species of birds including domestic poultry. Diseases caused by these viruses are severe and result in devastating economic impact due to high mortality and trade restrictions. In poultry, co-infections with both viruses are common, especially in endemic areas (Costa-Hurtado et al., Citation2015).
Influenza A viruses, members of the family Orthomyxoviridae, are segmented negative-sense ssRNA viruses. The avian influenza virus (AIV) is categorized into several subtypes based on its surface glycoproteins haemagglutinin (HA) and neuraminidase (Fouchier et al., Citation2005). The AIVs can also be classified into highly pathogenic avian influenza (HPAI) and low pathogenic avian influenza viruses (Swayne and Suarez, Citation2000) according to their pathogenicity in chickens; the HPAIVs induce systemic infections, particularly in chickens and turkeys, due to their furin-sensitive HA cleavage site (Alexander, Citation1993).
NDV, a prototype of avian paramyxovirus-1, causes a contagious notifiable disease, which is on the “list A” of the World Organization for Animal Health (Office International des Epizooties). The virus belongs to the genus Avulavirus within the family Paramyxoviridae (ICTV, Citation2012). The viral genome is linear, non-segmented, negative-sense ssRNA approximately 15 kb in size and contains six genes coding for nucleocaspid protein, phosphoprotein, matrix protein, fusion protein, haemagglutinin–neuraminidase and large polymerase protein (Czeglédi et al., Citation2006). Based on the phylogenetic topology of F gene sequences, NDV can be divided into class I and class II; class I is usually avirulent (Liu et al., Citation2009), while class II consists of 18 genotypes and includes both virulent and avirulent strains (Diel et al., Citation2012; Snoeck et al., Citation2013). According to its pathogenicity, NDV is categorized into five pathotypes: viscerotropic velogenic, neurotropic velogenic, mesogenic, lentogenic and asymptomatic enteric (Alexander, Citation2011).
The F protein cleavage site (F0) of NDV is considered the major determinant of virulence (Panda et al., Citation2004). In velogenic and mesogenic strains, a multi-basic amino acid (aa) motif, 112R/G/K-R-Q/K-K/R-R↓F117, at F0 is cleaved by ubiquitous intracellular proteases. The lentogenic strains, on the other hand, have a monobasic aa motif at F0, 112GR/K-Q-G-R↓L117, which can be cleaved by extracellular proteases found in the tissues of respiratory and intestinal tracts (Fujii et al., Citation1999). Other viral factors have also been involved in the determination of virus infectivity (Park et al., Citation2003).
Pigeon-origin NDV, also called pigeon paramyxovirus type-1 (PPMV-1), is an antigenic variant of avian paramyxovirus-1 and is known to infect pigeons, doves, wild birds and domestic poultry. The disease is similar to the nervous form of Newcastle disease (ND), with few obvious respiratory signs (Marlier and Vindevogel, Citation2006; Guo et al., Citation2013). PPMV-1 can be differentiated from classical NDV in several ways including haemagglutination inhibition test, monoclonal antibodies and restriction F gene cleavage site analysis. Usually, PPMV-1 isolates are classified, phylogenetically, as a distinct sub-genotype within genotype VI of class II (sub-genotype VIb) (Ujvári et al., Citation2003). PPMV-1 can infect chickens but not as severely as the velogenic strains of NDV (Werner et al., Citation1999; Abolnik et al., Citation2004). Historically, the first discovery of PPMV-1 was in 1978 in Iraq from diseased pigeons with encephalomyelitis (Tantawi et al., Citation1979). Four major panzootics of ND have been reported in different avian species (Miller and Koch, Citation2013). Of these, the third outbreak, mainly in pigeons and doves, was caused by PPMV-1 that originated from the Middle East (Iraq) in the late 1970s and spread rapidly to Europe (Kaleta et al., Citation1985). Now, PPMV-1 is endemic in domestic and feral pigeons in many areas of the world including the USA and Europe (Aldous et al., Citation2014).
In many countries, vaccination of racing pigeons against NDV is compulsory. However, there is no form of control in wild pigeons (Toro et al., Citation2005). That is why pigeons are implicated as carriers, which threaten the poultry industry (Carrasco et al., Citation2008). The situation in Egypt varies as most pigeon-rearing policies do not include routine vaccination against NDV. Recently, pigeons in Egypt have experienced several viral infections under natural conditions (Mansour et al., Citation2014; Elgendy et al., Citation2016; Rohaim et al., Citation2016). The purpose of this study is to investigate the outbreaks of nervous signs and deaths in pigeons in Egypt and to conduct molecular characterization of the isolated pathogens.
Materials and methods
Sample source
Between 2013 and 2015, pigeons in the Sharkia Province of Egypt experienced neurological signs, greenish diarrhoea, respiratory signs, conjunctivitis and death. Pigeons from 31 different houses (one symptomatic live pigeon per house) were submitted by the owners to the diagnostic laboratory. The number of birds per house ranged between 7 and 400, with an average of 133 pigeons per house. The age of the pigeons ranged between 15 days and 3 years.
Virus isolation and RNA extraction
Following clinical examination, the birds were euthanized for necropsy. Brain tissues were collected, homogenized into a 10% suspension using sterile phosphate-buffered saline (pH 7.2) and centrifuged at 2000 × g for 10 min. The supernatant fluid from each sample was inoculated into the allantoic sac of five, 9 to 11-day-old embryonated chicken eggs, which were then incubated at 37°C for 5 days (OIE, Citation2012). Subsequently, the allantoic fluids were harvested and tested for haemagglutination activity. Samples found negative were inoculated successively for another two blind passages. Viral RNA was extracted from haemagglutination-positive allantoic fluids using a Blood/Liquid Sample Total RNA Rapid Extraction Kit (Bioteke Corporation, Beijing, China) according to the manufacturer’s instructions.
Reverse transcription–polymerase chain reaction
The extracted RNA was screened using Matrix gene-based real-time reverse transcription–polymerase chain reaction (RT-PCR) targeting a conserved region of influenza A viruses using a one-step RT-PCR Kit (Qiagen, Valencia, CA, USA) and TaqMan fluorescein amidite-labelled hydrolysis probe (). The reaction was carried out in a total volume of 20 μl as described previously elsewhere (Spackman et al., Citation2002). Initially, the reverse transcription was done at 50°C for 30 min followed by PCR activation at 95°C for 15 min. After that, 45 amplification cycles of 94°C for 1 s and 60°C for 20 s were implemented for denaturation and primer annealing, respectively. After that, the samples with amplified M segments were subtyped for HA gene using a one-step RT-PCR Kit (Qiagen) and specific primers to cover the HA cleavage sites of the H5, H7 and H9 subtypes of AIV (). The samples were amplified for 35 cycles following the next protocol (heat denaturation at 94°C for 30 s, primer annealing at 52°C for 30 s and primer extension at 72°C for 45 s) proceeded by a two-step pre-cycle of 50°C for 30 min and 95°C for 15 min and followed by one cycle of final primer extension at 72°C for 10 min. Using a specific primer for the detection of NDV (), the same protocol was applied but with 94°C for 1 min for denaturation, 55°C for 1 min for primer annealing and 1 min for primer extension. The amplified products were visualized on 1.2% agarose gels.
Table 1. Oligonucleotide primers and probe used in the study.
Sanger sequencing
The viral PCR products were purified using a PCR purification kit (Qiagen). Purified DNA was sequenced separately using forward and reverse primers at the University of Minnesota Genomic Center. Consensus sequences were generated from the alignment of forward and reverse sequences using Sequencher software version 5.1 (http://genecodes.com/). Sequences obtained were compared with the existing database using the online Blast research tool (http://www.ncbi.nlm.nih.gov/).
Phylogenetic analysis
Comparative alignment was performed using the Clustal W method in MEGA 6.0 software. The per cent of nucleotide (nt%) and aa% identities was calculated using pairwise (p) distances. Phylogenetic trees were constructed using the neighbour-joining statistical method of MEGA 6.0 software with a topology of 1000 bootstrapping (Tamura et al., Citation2013).
nt sequence accession number
The AIV-HA gene and NDV-F gene nt sequences obtained in this study are available at GenBank under accession numbers KX580994 to KX580998 and KX580975 to KX580993, respectively.
Statistical analysis
The detection rate of influenza and paramyxo-viruses was calculated as the proportion of the samples in which infection was detected by RT-PCR. Both detection rate and 95% confidence interval (CI) were computed using WinPepi software, Version 11.65 (Abramson, Citation2011).
Results
The clinical findings
Neurological signs of torticollis, head tremors, leg and wing paralysis and incoordination were observed in 24 of the 31 tested pigeons (77.4%). Four birds showed respiratory signs (4/31) and 10 birds (10/31) showed conjunctivitis. All investigated pigeons exhibited greenish diarrhoea. In three of 31 houses, the mortality rate was ∼90%. Gross examination showed soft and friable brain tissues with congested meningeal blood vessels and/or haemorrhages. Necrotic foci on the pancreas were noticed in 2/31 pigeons (). The lungs were congested and/or haemorrhagic. Petechial haemorrhages on coronary fat were observed in 3/31 pigeons. A preliminary diagnosis of HPAI or NDV (PPMV-1) infection was made.
Table 2. Mortalities and clinical findings of influenza and paramyxo-viruses in pigeons in Sharkia Province, Egypt, during 2013–2015.
Virus isolation and haemagglutination
The infected embryonated chicken eggs manifested embryo mortality within 72 hours. Based on embryo lesions and HA, 24 of the 31 samples (77.4%) showed evidence of viral infection. The HA titre of various allantoic fluids ranged from 1:128 to 1:1024.
Detection of AIV and NDV (PPMV-1)
The HA-positive allantoic fluids were confirmed, by real time RT-PCR, to be AIV positive (61.3%; 19/31 pigeons) with 95% CI: 42.2–78.2%. Using RT-PCR, all positive samples were subtyped as HPAI-H5. Neither H7 nor H9 subtypes were detected. Meanwhile, when the same allantoic fluids were screened for NDV, 21/31 (67.8%) were found to be positive (95% CI 48.6–83.3%). The detection rate of AIV and NDV was explored according to four factors: season, age, breed of birds and housing system (). For AIV infections, the detection percentages ranged from 46.2% in winter to 100% in autumn. Also 81.8% of foreign breeds were infected. The highest AIV detection (70%) was recorded in birds of age range from 1 month to 1 year, while 80% of pigeons reared in the closed system were infected compared with the open system (52.4%). For NDV infections, the detection percentages ranged from 50% in autumn to 71.4% in summer, 72.7% of foreign breeds were infected, 83.3% of birds in the age range of ≤ 1 month were infected, as were 71.4% of those in the open rearing system. Furthermore, co-infection with both viruses was detected in 16 out of the 31 samples with a percentage of 51.6%. The co-infections appeared to predominate throughout all factors, especially with foreign breeds of pigeons (63.6%), between 1-month and 1-year of age (55%), in the closed system (60%) and in summer (71.4%) (). Further statistical analysis should be performed to describe the significance of these factors.
Figure 1. Detection of HPAIV, PPMV-1 and co-infection with both viruses in pigeons by season, age, breed and housing system using RT-PCR.
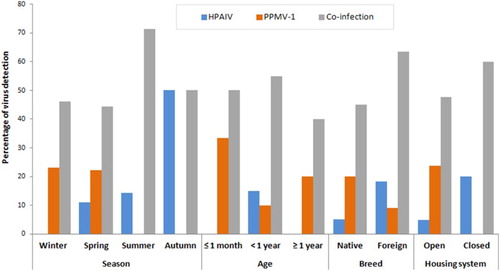
Table 3. Molecular detection of HPAIV and PPMV-1 in pigeons in Sharkia Province, Egypt, during 2013–2015.
Sequencing and phylogenetic analysis
The AIV sequences possessed multi-basic aas at the HA gene cleavage site (PQGEKRRKKR/GLF). The phylogenetic tree revealed that they belonged to clade 2.2.1.2. The HA gene sequence comparison showed that they shared identities of 99.6%–100% at the nt level and 100% at the aa level. They showed a high degree of identity with different Egyptian AIV isolates available in GenBank (data not shown).
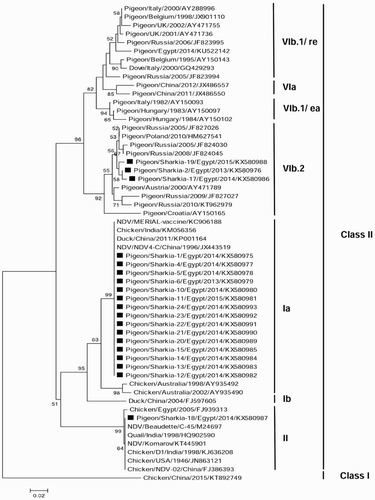
The phylogenetic tree was constructed using the neighbour-joining statistical method in MEGA 6.0 software based on the partial nt sequences of the NDV-F protein gene (). The study sequences (n = 19) belonged to NDV-class II; 15 were in genotype I (lineage 1) and clustered as sub-genotype Ia, three were in genotype VI (lineage 4) and clustered as sub-genotype VIb.2 and 1 isolate was in genotype II (lineage 2). The sequence analysis of the F protein cleavage site revealed that the genotype Ia, VIb.2 and II sequences carried the 112GKQGRL117, 112KRQKRF117and 112RRQKRF117 motifs, respectively. The overall identities between the study sequences were 81.5–100% at the nt level and 81.9–100% at the aa level. Genotype Ia sequences were 99.6–100% identical at the nt level and 100% identical at the aa level. The VIb.2 sequences showed identities of 98.5–99.5% and 98.5–100% at the nt and aa levels, respectively. The nt and aa identities of the study sequences were compared with those of other reference isolates from GenBank and listed in supplementary Tables 1–3.
Figure 2. The neighbour-joining phylogenetic tree of the Egyptian PPMV-1 isolates based on the partial nt sequences of F protein gene (635 bp). A Chinese NDV-Class I strain was selected as an out-group sequence. Bootstrap values of >50% are shown above the branches. The isolates of this study are marked with solid squares.
Discussion
HPAIV and NDV are two of the most common causes of disease outbreaks in poultry around the world. In Egypt, both diseases are enzootic and result in huge economic losses due to high morbidity and mortality rates, culling of infected and in-contact birds, loss of meat and eggs, cost of veterinary care and medications, governmental compensation of poultry farmers and imposing of trade restrictions. Pigeons are considered as a major threat for the transmission of both viruses to domestic poultry (Carrasco et al., Citation2008) because of their migratory nature, difficulties in vaccine application and, perhaps, being available in live bird markets, backyard houses and even in commercial poultry farms either intentionally or unintentionally. Also pigeons can easily be in contact with free-range birds. Feedstuff contaminated with faeces of NDV-infected pigeons is the main source of infection in many outbreaks (Toro et al., Citation2005; Ezema et al., Citation2009).
The present study focused on the outbreaks of influenza and paramyxovirus infections among clinically diseased pigeons. Variable mortalities (10–92.5%), in both single and mixed virus infections, were recorded. The ability of virus to cause a broad range of such variations may be attributed to co-infection with other pathogens, bird status and environmental factors (Aldous and Alexander, Citation2001). Furthermore, the lack of good hygiene and biosafety precautions, weak governmental efforts for enforcing preventive measures and inefficiently studied vaccination programmes are the main predisposing factors for such problems (ElMasry et al., Citation2015). In this study, the infected pigeons showed a clinical picture indicative of AIV and/or PPMV-1; pigeons showed tremors (20/24), torticollis (4/24), wing and leg paralysis (6/24) and greenish diarrhoea that are often seen (Barton et al., Citation1992; Marlier and Vindevogel, Citation2006). In PPMV-1 infections, respiratory signs were observed only in few naturally infected pigeons (Guo et al., Citation2013). Our results revealed that only four infected pigeons, either with HPAIV alone or in combination with PPMV-1, suffered from respiratory illness. Necrotic foci on the pancreas were seen in two pigeons infected with HPAIV.
In Egypt, AIVs started to infect pigeons naturally (Mansour et al., Citation2014), humans (Kayali et al., Citation2016) and other birds (Kayali et al., Citation2014). Over time, the viruses have evolved, mutated and changed their pathogenicity. The rate of AIV detection in our study (61.3%) is much more than that reported by Mansour et al. (Citation2014) and Elgendy et al. (Citation2016), indicating that the virus has disseminated in pigeons under natural conditions. The HPAIV clustered in group 2.2.1.2, which is currently predominant in Egypt.
Since its discovery in the Middle East, the PPMV-1 is still circulating around the world causing considerable economic losses (Aldous et al., Citation2014). In Egypt, the most dominant NDV genotypes are II, VI and VII (Radwan et al., Citation2013). In this study, the overall detection of pigeon-origin NDV was 67.8%. Genotypes I (79%), II (5.25%) and VI (15.75%) were detected in clinically diseased pigeons. The deduced aa sequence analysis revealed that they possessed the lentogenic motif 112GKQGRL117 at F0, which is characteristic of the avirulent NDV strains, and have high identity with the vaccine strain (KC906188) used in Egypt. Although 79% of our sequences were clustered with genotype I of NDV that includes the lentogenic and vaccine strains (Aldous et al., Citation2003), these isolates may have potential virulence in chickens by multiple passages (Kommers et al., Citation2001). Lentogenic strains of NDV can induce considerable losses when spread in the field by causing mild respiratory illness, which can be exaggerated, with other respiratory pathogens and/or environmental stressors. Besides, 10/15 of our genotype I isolates were identified from flocks with no history of previous NDV vaccination. Surprisingly, a high level of nervous manifestations was observed probably because of simultaneous infection with lentogenic-NDV and HPAIV subtype H5N1. The infection of 11/15 pigeons with HPAIV and lentogenic-NDV genotype I suggests that they may play a significant role in the epidemiology of both viruses in Egypt. Meanwhile, only one sequence was clustered in NDV-genotype II, carrying the velogenic motif 112RRQKRF117, which is known to be common among chickens in Egypt. This may be due to easy intermixing of pigeons and chickens.
Three sequences were clustered with the poorly studied group, VIb.2, which is strictly defined as PPMV-1 (Ujvári et al., Citation2003). These sequences carried the velogenic motif 112KRQKRF117. As a group, VIb.2 contains the isolates of Croatia, Austria, Poland and Russia and has a relatively stable geographical distribution in populations of feral pigeons (Pchelkina et al., Citation2012). Most recently, Rohaim et al. (Citation2016) reported the presence of PPMV-1 genotype VIb.1, a classical pigeon lineage, which clustered with recent European lineage (EU/re) of VIb.1 (Ujvári et al., Citation2003; Pchelkina et al., Citation2012). This indicates that there are several sub-genotypes currently circulating in pigeons in Egypt. It is possible that they might have spread from Europe to Egypt through migration and/or importation of pigeons.
Co-infection of poultry with more than one viral and/or bacterial agent is common and results in a complicated clinical picture. In co-infected birds, the first virus may influence the tropism, replication strategy and immune responses of the second virus, which is defined as viral interference (DaPalma et al., Citation2010). This may lead to incomplete diagnosis of causative agents (El Zowalaty et al., Citation2011). In Egypt, Hassan et al., (Citation2016) reported that chickens were co-infected with several respiratory viruses including AIV-H5, AIV-H9, NDV and infectious bronchitis virus. Our study reported the co-infection of pigeons with AIV-H5 and NDV (51.6%). Other parameters such as the strains of viruses involved, challenge titre of the viruses and timing of co-infections should be taken into consideration because they will affect the pattern of infections and the pathogenesis of both viruses (Costa-Hurtado et al., Citation2015).
In conclusion, this study reported, for the first time, the outbreaks of pigeon-origin NDV (PPMV-1) and HPAI-H5N1 striking Egypt between 2013 and 2015 on a large scale (31 pigeon houses) compared with previous studies (Mansour et al., Citation2014; Elgendy et al., Citation2016; Rohaim et al., Citation2016). We characterized the circulating PPMV-1 genotypes as Ia, II and VIb.2, suggesting the co-circulation of different NDV genotypes among pigeons in Egypt. It could be concluded that the concurrent mixed AIV and NDV infections are prevalent in pigeons of Egypt. The results obtained may explain the vaccination failure reported in commercial poultry farms in Egypt. Thus, our study could be considered as a wake-up call due to the possibility of the emergence of new viruses, transmission to other poultry species, particularly chicken, and occurrence of uncontrolled epidemic outbreaks in the future. Active surveillance of both viruses in pigeons is necessary to understand their dissemination in Egypt. The results of this study may be helpful in the selection of the optimum vaccination strategy for the prevention and control of future outbreaks.
Supplemental data
Download MS Word (76.5 KB)Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Abolnik, C., Horner, R.F., Maharaj, R. & Viljoen, G.J. (2004). Characterization of a pigeon paramyxovirus (PPMV-1) isolated from chickens in South Africa. The Onderstepoort Journal of Veterinary Research, 71, 157–160. doi: 10.4102/ojvr.v71i2.278
- Abramson, J.H. (2011). WinPepi updated: computer programs for epidemiologists, and their teaching potential. Epidemiologic Perspectives and Innovations, 8, 1. doi: 10.1186/1742-5573-8-1
- Aldous, E.W. & Alexander, D.J. (2001). Detection and differentiation of Newcastle disease virus (avian paramyxovirus type 1). Avian Pathology, 30, 117–128. doi: 10.1080/03079450120044515
- Aldous, E.W., Fuller, C.M., Ridgeon, J.H., Irvine, R.M., Alexander, D.J. & Brown, I.H. (2014). The evolution of pigeon paramyxovirus type 1 (PPMV-1) in Great Britain: a molecular epidemiological study. Transboundary and Emerging Diseases, 61, 134–139. doi: 10.1111/tbed.12006
- Aldous, E.W., Mynn, J.K., Banks, J. & Alexander, D.J. (2003). A molecular epidemiological study of avian paramyxovirus type 1 (Newcastle disease virus) isolates by phylogenetic analysis of a partial nucleotide sequence of the fusion protein gene. Avian Pathology, 32, 239–256. doi: 10.1080/030794503100009783
- Alexander, D.J. (1993). Orthomyxoviruses. In J.B. McFerran & M.S. McNulty (Eds.), Virus Infections of Birds (pp. 287–316). London: Elsevier Science.
- Alexander, D.J. (2011). Newcastle disease in the European union 2000 to 2009. Avian Pathology, 40, 547–558. doi: 10.1080/03079457.2011.618823
- Barton, J.T., Bickford, A.A., Cooper, G.L., Charlton, B.R. & Cardona, C.J. (1992). Avian paramyxovirus type 1 infections in racing pigeons in California. I. Clinical signs, pathology, and serology. Avian Diseases, 36, 463–468. doi: 10.2307/1591531
- Carrasco, A.D.O.T., Seki, M.C., de Freitas Raso, T., Paulillo, A.C. & Pinto, A.A. (2008). Experimental infection of Newcastle disease virus in pigeons (Columba livia): humoral antibody response, contact transmission and viral genome shedding. Veterinary Microbiology, 129, 89–96. doi: 10.1016/j.vetmic.2007.11.012
- Chakrabarti, S., King, D.J., Afonso, C., Swayne, D., Cardona, C.J., Kuney, D.R. & Gerry, A.C. (2007). Detection and isolation of exotic Newcastle disease virus from field-collected flies. Journal of Medical Entomology, 44, 840–844. doi: 10.1093/jmedent/44.5.840
- Costa-Hurtado, M., Afonso, C.L., Miller, P.J., Shepherd, E., Cha, R.M., Smith, D., Spackman, E., Kapczynski, D.R., Suarez, D.L., Swayne, D.E. & Pantin-Jackwood, M.J. (2015). Previous infection with virulent strains of Newcastle disease virus reduces highly pathogenic avian influenza virus replication, disease, and mortality in chickens. Veterinary Research, 46, 97. doi: 10.1186/s13567-015-0237-5
- Czeglédi, A., Ujvári, D., Somogyi, E., Wehmann, E., Werner, O. & Lomniczi, B. (2006). Third genome size category of avian paramyxovirus serotype 1 (Newcastle disease virus) and evolutionary implications. Virus Research, 120, 36–48. doi: 10.1016/j.virusres.2005.11.009
- DaPalma, T., Doonan, B.P., Trager, N.M. & Kasman, L.M. (2010). A systematic approach to virus–virus interactions. Virus Research, 149, 1–9. doi: 10.1016/j.virusres.2010.01.002
- Diel, D.G., da Silva, L.H., Liu, H., Wang, Z., Miller, P.J. & Afonso, C.L. (2012). Genetic diversity of avian paramyxovirus type 1: proposal for a unified nomenclature and classification system of Newcastle disease virus genotypes. Infection, Genetics and Evolution, 12, 1770–1779. doi: 10.1016/j.meegid.2012.07.012
- Elgendy, E.M., Watanabe, Y., Daidoji, T., Arai, Y., Ikuta, K., Ibrahim, M.S. & Nakaya, T. (2016). Genetic characterization of highly pathogenic avian influenza H5N1 viruses isolated from naturally infected pigeons in Egypt. Virus Genes, 52, 867–871. doi: 10.1007/s11262-016-1369-z
- ElMasry, I., Elshiekh, H., Abdlenabi, A., Saad, A., Arafa, A., Fasina, F.O., Lubroth, J. & Jobre, Y.M. (2015). Avian influenza H5N1 surveillance and its dynamics in poultry in live bird markets, Egypt. Transboundary and Emerging Diseases, 64, 805–814.
- El Zowalaty, M.E., Chander, Y., Redig, P.T., El Latif, H.K.A., El Sayed, M.A. & Goyal, S.M. (2011). Selective isolation of avian influenza virus (AIV) from cloacal samples containing AIV and Newcastle disease virus. Journal of Veterinary Diagnostic Investigation, 23, 330–332. doi: 10.1177/104063871102300222
- Ezema, W.S., Okoye, J.O.A. & Nwanta, J.A. (2009). LaSota vaccination may not protect against the lesions of velogenic Newcastle disease in chickens. Tropical Animal Health and Production, 41, 477–484. doi: 10.1007/s11250-008-9210-x
- Fouchier, R.A., Munster, V., Wallensten, A., Bestebroer, T.M., Herfst, S., Smith, D., Rimmelzwaan, G.F., Olsen, B. & Osterhaus, A.D. (2005). Characterization of a novel influenza A virus hemagglutinin subtype (H16) obtained from black-headed gulls. Journal of Virology, 79, 2814–2822. doi: 10.1128/JVI.79.5.2814-2822.2005
- Fujii, Y., Sakaguchi, T., Kiyotani, K. & Yoshida, T. (1999). Comparison of substrate specificities against the fusion glycoprotein of virulent Newcastle disease virus between a chick embryo fibroblast processing protease and mammalian subtilisin-like proteases. Microbiology and Immunology, 43, 133–140. doi: 10.1111/j.1348-0421.1999.tb02384.x
- Guo, H., Liu, X., Han, Z., Shao, Y., Chen, J., Zhao, S., Kong, X. & Liu, S. (2013). Phylogenetic analysis and comparison of eight strains of pigeon paramyxovirus type 1 (PPMV-1) isolated in China between 2010 and 2012. Archives of Virology, 158, 1121–1131. doi: 10.1007/s00705-012-1572-8
- Hassan, K.E., Shany, S.A., Ali, A., Dahshan, A.H.M., Azza, A. & El-Kady, M.F. (2016). Prevalence of avian respiratory viruses in broiler flocks in Egypt. Poultry Science, 95, 1271–1280. doi: 10.3382/ps/pew068
- ICTV. (2012). Virus taxonomy: 2012 release. London, UK.
- Kaleta, E.F., Alexander, D.J. & Russell, P.H. (1985). The first isolation of the avian PMV-1 virus responsible for the current panzootic in pigeons? Avian Pathology, 14, 553–557. doi: 10.1080/03079458508436258
- Kayali, G., Kandeil, A., El-Shesheny, R., Kayed, A.S., Gomaa, M.M., Maatouq, A.M., Shehata, M.M., Moatasim, Y., Bagato, O., Cai, Z. & Rubrum, A. (2014). Active surveillance for avian influenza virus, Egypt, 2010–2012. Emerging Infectious Diseases, 20, 542–551. doi: 10.3201/eid2004.131295
- Kayali, G., Kandeil, A., El-Shesheny, R., Kayed, A.S., Maatouq, A.M., Cai, Z., McKenzie, P.P., Webby, R.J., El Refaey, S., Kandeel, A. & Ali, M.A. (2016). Avian influenza A (H5N1) virus in Egypt. Emerging Infectious Diseases, 22, 379–388. doi: 10.3201/eid2203.150593
- Kommers, G.D., King, D.J., Seal, B.S. & Brown, C.C. (2001). Virulence of pigeon-origin Newcastle disease virus isolates for domestic chickens. Avian Diseases, 45, 906–921. doi: 10.2307/1592870
- Liu, X., Wang, X., Wu, S., Hu, S., Peng, Y., Xue, F. & Liu, X. (2009). Surveillance for avirulent Newcastle disease viruses in domestic ducks (Anas platyrhynchos and Cairina moschata) at live bird markets in Eastern China and characterization of the viruses isolated. Avian Pathology, 38, 377–391. doi: 10.1080/03079450903183637
- Mansour, S.M., ElBakrey, R.M., Ali, H., Knudsen, D.E. & Eid, A.A. (2014). Natural infection with highly pathogenic avian influenza virus H5N1 in domestic pigeons (Columba livia) in Egypt. Avian Pathology, 43, 319–324. doi: 10.1080/03079457.2014.926002
- OIE. (2012). Manual of Diagnostic Tests and Vaccines for Terrestrial Animals (chapter 2.3.14).
- Marlier, D. & Vindevogel, H. (2006). Viral infections in pigeons. The Veterinary Journal, 172, 40–51. doi: 10.1016/j.tvjl.2005.02.026
- Miller, P.J. & Koch, G. (2013). Newcastle disease. In D.E. Swayne, J.R. Glisson, L.R. McDougald, L.K. Nolan, D.L. Suarez & V. Nair (Eds.). Diseases of poultry 13th edn (pp. 89–107). Ames, IA: Wiley-Blackwell.
- Panda, A., Huang, Z., Elankumaran, S., Rockemann, D.D. & Samal, S.K. (2004). Role of fusion protein cleavage site in the virulence of Newcastle disease virus. Microbial Pathogenesis, 36, 1–10. doi: 10.1016/j.micpath.2003.07.003
- Park, M.S., Shaw, M.L., Munoz-Jordan, J., Cros, J.F., Nakaya, T., Bouvier, N., Palese, P., García-Sastre, A. & Basler, C.F. (2003). Newcastle disease virus (NDV)-based assay demonstrates interferon-antagonist activity for the NDV V protein and the Nipah virus V, W, and C proteins. Journal of Virology, 77, 1501–1511. doi: 10.1128/JVI.77.2.1501-1511.2003
- Pchelkina, I.P., Manin, T.B., Kolosov, S.N., Starov, S.K., Andriyasov, A.V., Chvala, I.A., Drygin, V.V., Yu, Q., Miller, P.J. & Suarez, D.L. (2012). Characteristics of pigeon paramyxovirus serotype-1 isolates (PPMV-1) from the Russian Federation from 2001 to 2009. Avian Diseases, 57, 2–7. doi: 10.1637/10246-051112-Reg.1
- Radwan, M.M., Darwish, S.F., El-Sabagh, I.M., El-Sanousi, A.A. & Shalaby, M.A. (2013). Isolation and molecular characterization of Newcastle disease virus genotypes II and VIId in Egypt between 2011 and 2012. Virus Genes, 47, 311–316. doi: 10.1007/s11262-013-0950-y
- Rohaim, M.A., El Naggar, R.F., Helal, A.M., Hussein, H.A. & LeBlanc, N. (2016). Genetic characterization of pigeon paramyxovirus type 1 in Egypt. British Journal of Virology, 3, 27–32. doi: 10.17582/journal.bjv/2016.3.2.27.32
- Snoeck, C.J., Owoade, A.A., Couacy-Hymann, E., Alkali, B.R., Okwen, M.P., Adeyanju, A.T., Komoyo, G.F., Nakouné, E., Le Faou, A. & Muller, C.P. (2013). High genetic diversity of Newcastle disease virus in poultry in West and Central Africa: cocirculation of genotype XIV and newly defined genotypes XVII and XVIII. Journal of Clinical Microbiology, 51, 2250–2260. doi: 10.1128/JCM.00684-13
- Spackman, E., Senne, D.A., Myers, T.J., Bulaga, L.L., Garber, L.P., Perdue, M.L., Lohman, K., Daum, L.T. & Suarez, D.L. (2002). Development of a real-time reverse transcriptase PCR assay for type A influenza virus and the avian H5 and H7 hemagglutinin subtypes. Journal of Clinical Microbiology, 40, 3256–3260. doi: 10.1128/JCM.40.9.3256-3260.2002
- Swayne, D.E. & Suarez, D.L. (2000). Highly pathogenic avian influenza. Revue Scientifique et Technique-office International des Epizooties, 19, 463–482. doi: 10.20506/rst.19.2.1230
- Tamura, K., Stecher, G., Peterson, D., Filipski, A. & Kumar, S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Molecular Biology and Evolution, 30, 2725–2729. doi: 10.1093/molbev/mst197
- Tantawi, H.H., Al Falluji, M.M. & Al Sheikhly, F. (1979). Viral encephalomyelitis of pigeons: identification and characterization of the virus. Avian Diseases, 23, 785–793. doi: 10.2307/1589594
- Toro, H., Hoerr, F.J., Farmer, K., Dykstra, C.C., Roberts, S.R. & Perdue, M. (2005). Pigeon paramyxovirus: association with common avian pathogens in chickens and serologic survey in wild birds. Avian Diseases, 49, 92–98. doi: 10.1637/7268-083104R1
- Tsukamoto, K., Ashizawa, H., Nakanishi, K., Kaji, N., Suzuki, K., Okamatsu, M., Yamaguchi, S. & Mase, M. (2008). Subtyping of avian influenza viruses H1 to H15 on the basis of hemagglutinin genes by PCR assay and molecular determination of pathogenic potential. Journal of Clinical Microbiology, 46, 3048–3055. doi: 10.1128/JCM.02386-07
- Ujvári, D., Kaleta, E.F., Werner, O., Savić, V., Nagy, É, Czifra, G. & Lomniczi, B. (2003). Phylogenetic analysis reveals extensive evolution of avian paramyxovirus type 1 strains of pigeons (Columba livia) and suggests multiple species transmission. Virus Research, 96, 63–73. doi: 10.1016/S0168-1702(03)00173-4
- Werner, O., Römer-Oberdörfer, A., Köllner, B., Manvell, R.J. & Alexander, D.J. (1999). Characterization of avian paramyxovirus type 1 strains isolated in Germany during 1992 to 1996. Avian Pathology, 28, 79–88. doi: 10.1080/03079459995082
