ABSTRACT
A quantitative Polymerase Chain Reaction (qPCR) for the seven chicken Eimeria spp. was modified and validated for direct use on fresh droppings. The analytical specificity of the qPCR on droppings was 100%. Its analytical sensitivity (non-sporulated oocysts/g droppings) was 41 for E. acervulina, ≤2900 for E. brunetti, 710 for E. praecox, 1500 for E. necatrix, 190 for E. tenella, 640 for E. maxima, and 1100 for E. mitis. Field validation of the qPCR was done using droppings with non-sporulated oocysts from 19 broiler flocks. To reduce the number of qPCR tests five grams of each pooled sample (consisting of ten fresh droppings) per time point were blended into one mixed sample. Comparison of the oocysts per gram (OPG)-counting method with the qPCR using pooled samples (n = 1180) yielded a Pearson’s correlation coefficient of 0.78 (95% CI: 0.76–0.80) and a Pearson’s correlation coefficient of 0.76 (95% CI: 0.70–0.81) using mixed samples (n = 236). Comparison of the average of the OPG-counts of the five pooled samples with the mixed sample per time point (n = 236) showed a Pearson’s correlation coefficient (R) of 0.94 (95% CI: 0.92–0.95) for the OPG-counting method and 0.87 (95% CI: 0.84–0.90) for the qPCR. This indicates that mixed samples are practically equivalent to the mean of five pooled samples. The good correlation between the OPG-counting method and the qPCR was further confirmed by the visual agreement between the total oocyst/g shedding patterns measured with both techniques in the 19 broiler flocks using the mixed samples.
Introduction
Coccidiosis is a disease with a very significant economic impact in poultry farming, which is caused by intestinal protozoa belonging to the subclass Coccidia. Their life cycle starts with the ingestion of sporulated oocysts, which can be found in the poultry house environment. During infection the mucosal surfaces of the gastrointestinal tract can be damaged by the intracellular replication of the parasites (McDougald & Fitz-Coy, Citation2013). Depending on the severity of the infection, the virulence of the Eimeria species involved and the occurrence of co-infecting pathogens, the manifestation of the disease may vary from impaired absorption of nutrients to haemorrhagic or necrotic enteritis, leading to starvation and death. Direct economic damage is caused by reduced growth, increased feed conversion, and higher mortality, while the indirect economic impact is inflicted by costs associated with preventive and therapeutic intervention strategies. The global annual costs in commercial poultry have been estimated to be more than 2 billion euros (Shirley et al., Citation2005).
Traditionally, the diagnosis of coccidiosis is made by assessing the occurrence and severity of macroscopic lesions in the intestinal tract during post-mortem examination (Johnson & Reid, Citation1970), often aided by microscopic examination of scrapings of the intestinal mucosa. Additionally, analysis of the number of oocysts/g droppings (oocysts per gram (OPG)-counting method) using a salt flotation technique (Hodgson, Citation1970) may be performed. Although this procedure may suffice for basic diagnostic work, it has a number of drawbacks: (1) birds must be killed; (2) the number of birds examined is generally low; (3) birds selected for post-mortem examination may not represent the disease stage in the flock; (4) macroscopic lesions are not always explicit; and (5) microscopic identification of oocysts is not unambiguous either (Haug et al., Citation2008). Mentioned shortcomings could be circumvented by a quantitative Polymerase Chain Reaction (qPCR) technique able to detect and identify all seven chicken Eimeria spp. in fresh dropping samples.
Vrba et al. (Citation2010) described quantitative PCR assays for all seven chicken Eimeria spp. based on non-polymorphic SCARs (sequence characterized amplification regions). A great advantage of their method was the lack of cross-reactivity between Eimeria spp., which often occurred using the PCR described by Morgan et al. (Citation2009) (non-published data). In the present study the former PCRs were combined as three multiplex Eimeria spp. qPCR assays, while the extraction method was changed in order to be able to analyse fresh droppings directly.
The three multiplex Eimeria spp. qPCRs were validated by comparing the qPCR results with OPG-counts using: (1) dropping samples of Specified Pathogen Free (SPF) broilers inoculated with Eimeria spp.; (2) spiked dropping samples from SPF broilers; and (3) fresh dropping samples obtained from 19 commercial broiler flocks.
Materials and methods
Eimeria spp. reference strains
E. acervulina (Weybridge), E. maxima (Weybridge) and E. tenella (Houghton) strains were obtained from the Animal Health Veterinary Laboratories Agency (AHVLA, Weybridge, UK). The E. acervulina strain was provided in 1990, while the E. maxima and E. tenella strains were obtained in 2000. Since then, these strains have been rejuvenated approximately every half year at GD. E. necatrix, E. brunetti and E. praecox were kindly provided by Intervet Innovation GmbH (Schwabenheim, Germany, a subsidiary of Merck & Co., Inc., Whitehouse Station, NJ, USA). These isolates originated from the Houghton Poultry Research Station (HPRS, Houghton, UK). E. mitis was obtained from a Dutch rearing layer flock. All Eimeria spp. had been cloned by single oocyst isolation and the purity was assessed during propagation in SPF broiler chickens as described by Peek and Landman (Citation2003). Strains were considered to be pure if: only macroscopic lesions of the corresponding Eimeria spp. were found during post-mortem examination of inoculated chickens and oocysts in intestinal mucosal scrapings showed uniform morphology in agreement with the corresponding species (McDougald & Fitz-Coy, Citation2013).
SPF broilers were inoculated with oocysts of either one of the cloned Eimeria spp. at 8 days of age. Droppings of broilers inoculated with E. acervulina, E. praecox or E. mitis were collected from day 4 until day 6 post inoculation (p.i.), droppings of broilers that were given E. maxima, E. brunetti or E. tenella from day 5 until 7 p.i. and droppings of broilers inoculated with E. necatrix from day 6 until 8 p.i. Collected droppings were thoroughly homogenized and the number of non-sporulated oocysts/g was determined in triplicate as described by Peek and Landman (Citation2003). The obtained results are given in . Dropping samples containing these oocysts were used for the calibration and assessment of the detection limit of the qPCR assays.
Table 1. Number of non-sporulated oocysts/g droppings (OPG-counts) of samples originating from SPF broilers inoculated with either one of the seven chicken Eimeria spp. reference strains determined in triplicate.
Primers, probes and DNA extraction
Primers and probes as described previously (Vrba et al., Citation2010) were used.
The DNA extraction was performed with the NucleoSpin®Soil kit (Bioké, Leiden, The Netherlands). It includes a mechanical lysis step for the disruption of the oocyst walls and a very strong lysis buffer.
Briefly, 100 mg droppings were put into the NucleoSpin®Soil Bead Tube, and 700 µl buffer SL2 and 150 µl Enhancer SX were added. Then, the tube was homogenized 3 × 5 min in a bead-beater with 5 min intervals, during which it was placed in an ice bath to prevent overheating. After homogenization, the content was centrifuged (2 min at 11,000×g) and 500 µl of the supernatant was collected. A volume of 150 µl buffer SL3 was added to the supernatant. The mixture was then homogenized on a vortex mixer and put on ice (5 min) and thereafter centrifuged (1 min at 11,000×g). The supernatant was then transferred to a Nucleospin®Inhibitor Removal Column and centrifuged (1 min at 11,000×g). Thereafter, 250 µl buffer SB was added and homogenized on the vortex mixer (5 sec). The mixture was placed on the Nucleospin®Soil Column and centrifuged (1 min at 11,000×g). After that the column was washed four times with 500 µl buffer SB, 550 µl buffer SW1, 700 µl buffer SW and with 700 µl SW2, respectively. Between each washing the column was centrifuged (30 sec at 11,000×g). After the last washing the column was dried by centrifugation (2 min at 11,000×g) and the DNA was eluted with 30 µl buffer SE (1 min at room temperature) and centrifuged (11,000×g).
Multiplex Eimeria spp. qPCRs
Three multiplex Eimeria spp. qPCRs, further denoted as qPCR, were made by mixing primers and combining probes with different fluorescent labels. Herewith the number of Eimeria spp. qPCRs for the detection of the seven chicken Eimeria spp. in one dropping sample was reduced from seven to three. The following combinations were made: E. acervulina and E. brunetti (mix 1), E. maxima and E. mitis (mix 2), and E. necatrix, E. praecox and E. tenella (mix 3).
Briefly, per sample 0.5 µl containing 10 pmol of each primer and 0.5 µl containing 2.6 pmol of each probe (Biolegio, Nijmegen, the Netherlands), 2 µl Quantifast Probe PCR mix (Qiagen, Hilden, Germany), 0.4 µl ROX and 2 µl BSA were mixed with water to a total volume of 17 µl. Per PCR reaction 3 µl DNA was added. The thermal cycler protocol was as follows: initial denaturation and enzyme activation for 15 min at 95°C followed by 45 cycles of 15 sec at 95°C denaturation and 30 sec at 60°C primer annealing and extension. Real-time PCR was performed on the ABI7500 fast system (Life Technologies, Foster City, CA, USA). Raw data were analysed with the Applied-Biosystem SDS software (Life Technologies). Fluorescence curves were analysed on a linear scale. Threshold was set at 0.2 for E. acervulina, E. brunetti and E. tenella, and at 0.1 for E. maxima, E. mitis, E. necatrix and E. praecox. Baseline correction was based on cycles 3–20 for E. tenella, 3–22 for E. acervulina, E. brunetti, E. maxima, E. mitis, and on 3–24 for E. necatrix and E. praecox.
In order to determine the PCR efficiency of the multiplex assays 10-fold serial dilutions of plasmid DNA containing the PCR target sequence from a single Eimeria species were prepared in nuclease-free water. Concentrations of the 10-fold dilutions ranged from 1.3 × 101–1.3 × 105 plasmid copies/PCR reaction. For each Eimeria species the different dilutions were tested in quadruplicate in the corresponding multiplex PCR. The PCR efficiency for individual species tested in the multiplex PCR was expressed as 10(−1/slope).
In vitro validation
Analytical specificity
The analytical specificity of the qPCR is defined as its ability to detect all strains of the species for which it has been designed (inclusivity) and its ability to exclude related organisms (i.e. to demonstrate exclusivity) (OIE, Citation2017a).
The analytical specificity of the qPCR was assessed using fresh droppings collected from each of 30 coccidiosis-free SPF birds and 30 dropping samples from the same birds spiked heterologously with Eimeria spp. (see analytical sensitivity). Moreover, in silico specificity testing of the primers and probes used in this assay was also performed by comparing their sequences to those of two other chicken parasites that also belong to the phylum Apicomplexa, namely Cryptosporidium spp. and Isospora spp., using BLAST search in Genbank. No significant homologies were found with any of the available sequences.
Analytical sensitivity
The analytical sensitivity is defined as the limit of detection of the assay and was determined using a dilution-to-extinction experiment (OIE, Citation2017b). Hereto, coccidiosis negative droppings of SPF birds were spiked with a decreasing number of oocysts of each of the Eimeria spp. reference strains. This was done by preparing 10-fold dilutions. However, 5-fold and 2-fold dilutions were made whenever droppings with high concentrations of oocysts per g were not obtained. In order to reliably determine the analytical sensitivity for each species investigated, a sufficient number of samples containing different concentrations of oocysts is needed.
The 10-fold dilution series for E. acervulina was prepared by adding 200 mg of droppings containing 4.1 × 106 non-sporulated oocysts (undiluted sample) to 1.8 gram of SPF droppings, which was followed by thorough mixing. This yielded dilution 1 with a concentration of 4.1 × 105 oocysts/g droppings. From this dilution 200 mg was added to 1.8 g of SPF droppings to obtain dilution 2 with a concentration of 4.1 × 104 oocysts/g droppings. Likewise, dilution 3, 4, 5, and 6 were prepared. The obtained concentrations for E. acervulina ranged from 4.1 × 106 to 4.1 × 100. Also for E. praecox 10-fold dilutions were made similarly, resulting in concentrations ranging from 7.1 × 105 to 7.1 × 100 oocysts/g droppings. Five-fold dilutions were made for E. maxima, E. mitis, and E. tenella. Hereto, 400 mg of droppings containing non-sporulated oocysts (undiluted sample) of E. maxima, E. mitis, or E. tenella was added to 1.6 g of SPF droppings, which was followed by thorough mixing (dilution 1). Dilution 2 was prepared by adding 400 mg of dilution 1 to 1.6 g of SPF droppings and so forth. The obtained concentrations ranged from 4.0 × 105 to 1.3 × 102 oocysts/g droppings for E. maxima, 1.4 × 105 to 4.3 × 101 oocysts/g for E. mitis and 1.3 × 105 to 3.8 × 101 for E. tenella. Finally, for E. brunetti and E. necatrix 2-fold dilutions were used. Dilution 1 was prepared by adding 1 g of droppings containing non-sporulated oocysts (undiluted sample) of E. brunetti or E. necatrix to 1 g of SPF droppings, followed by thorough mixing. Dilution 2 was prepared by adding 1 g of dilution 1 to 1 g of SPF droppings and so on. The concentrations obtained ranged from 9.3 × 104 to 2.9 × 103 oocysts/g droppings for E. brunetti and from 1.2 × 104 to 3.8 × 102 oocysts/g for E. necatrix.
The concentration series were tested in two-fold on three different days. The detection limit was the lowest concentration of oocysts that was detected in at least 50% of samples.
The detection limit of the OPG-counting technique used is 333 oocysts/g droppings, which was determined based on the following. Three grams of homogenized droppings were suspended in 30 ml NaCl solution (density = 1.1 g/ml) and vortexed (15 s). Thereafter, one ml of this suspension was transferred to a tube containing 9 ml NaCl solution (density = 1.2 g/ml). Immediately (that is, before allowing the oocysts come afloat) after vortexing (15 s), a McMaster counting chamber was filled with 0.3 ml (two chambers with a volume of 0.15 ml each) of the latter suspension. Considering the dilution steps, the presence of one oocyst in the counting chamber implies that 333 oocysts/g droppings are present.
Inter-assay repeatability
The inter-assay repeatability (within-laboratory reproducibility) of the qPCR is a measure of agreement between results (between runs in the same laboratory) (OIE, Citation2017a). It was assessed using three spiked samples for each Eimeria spp. with low, intermediate and high numbers of oocysts. Samples were analysed six times on different days. The inter-assay repeatability of the qPCR was expressed as a percentage coefficient of variation (CV) and calculated by dividing the standard deviation of Ct-values by the mean Ct-value × 100.
Field validation
Broiler flocks and samples of droppings
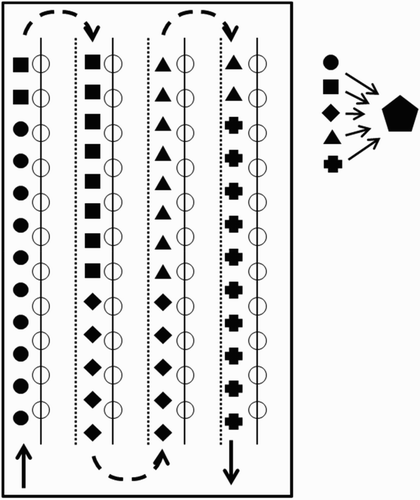
Samples of fresh droppings were collected from 19 broiler flocks kept on four different farms during successive grow-outs. On each farm, the same house was monitored during the study. Two farms belonged to feed mill A, and the others to feed mill B. Flocks were sampled three times a week (Monday, Wednesday, and Friday) starting at 14 days of age until the end of the grow-out period (approximately 42 days of age). Fifty fresh droppings (Hodgson, Citation1970), 10 (= one pooled sample) per polypropylene container of 120 ml (VWR reference code 216-0861, VWR International, Amsterdam, the Netherlands) were taken evenly distributed along four lanes in the broiler house per sampling (). The containers with samples of droppings were immediately stored in a refrigerator at 4–6°C to avoid sporulation in order to reduce the impact of sporulation on genome number per oocyst, attempting to improve the correlation between OPG and genome equivalents. After three consecutive samplings, the pooled samples were sent to GD overnight unrefrigerated. Upon arrival, containers were stored refrigerated again to avoid sporulation. After stirring pooled samples thoroughly, 5 g droppings were collected from each of the five pooled samples belonging to the same sampling, and blended into one mixed sample (). A total of 1416 samples (1180 pooled and 236 mixed) were obtained and analysed with the OPG-counting technique and the qPCR. Results of the OPG-counts were expressed as number of oocysts/g droppings and those of the qPCR as the sum of the number of oocyst equivalents/g droppings per Eimeria spp.
Figure 1. Route followed for the collection of samples of fresh droppings in the broiler houses. At each sampling, 50 fresh droppings were gathered evenly distributed along the walkway. The circles, squares, diamonds, triangles, and crosses illustrate the locations where a fresh dropping was taken. Pools of ten consecutively collected droppings were made (ten identical symbols represent one pool). Five g of each pooled sample was taken and blended to obtain one mixed sample (pentagon) at GD. Arrow = walking direction, continuous line with circles = line with feeding pans, dotted line = drinking lines.
Comparison of pooled and mixed samples
The mean number of oocyst/g droppings of the OPG-counting method and the mean number of oocyst equivalents/g droppings of the qPCR of the pooled samples were compared with the corresponding results of the mixed samples (n = 236). The results of the comparisons were expressed as Pearson’s correlation coefficient (Stata Corporation, Citation2011). This was done to assess whether mixed samples could substitute pooled samples for monitoring coccidiosis in the field.
Comparison of OPG-counts with the qPCRs
The results of the OPG-counting technique were compared with those of the qPCR for both pooled samples (n = 1180) and mixed samples (n = 236). The results of the comparisons were expressed as Pearson’s correlation coefficient (Stata Corporation, Citation2011).
Ethics
The study was approved by the Institutional Animal Experimental Committee, DEC-Consult Foundation, according to Dutch law on experimental animals (Wet op de dierproeven).
Results
Multiplex Eimeria spp. qPCRs.
PCR efficiencies for the Eimeria spp. tested in the multiplex PCRs were: 1.99 (sd = 0.06) for E. acervulina, 1.98 (sd = 0.06) for E. brunetti, 2.00 (sd = 0.08) for E. maxima, 2.00 (sd = 0.01) for E. mitis, 2.00 (sd = 0.02) for E. praecox, 2.00 (sd = 0.06) for E. necatrix, and 1.99 (sd = 0.05) for E. tenella.
In vitro validation
Analytical specificity
Exclusivity for the specific species was demonstrated for the primers and probes described by Vrba et al. (Citation2010) and no cross-reaction with other species was observed. This resulted in an analytical specificity of 100%. The results are shown in .
Table 2. Target loci, reporter dye and quencher, analytical specificity, detection limit, and reproducibility of the three multiplex Eimeria spp.
Analytical sensitivity
The detection limit of the qPCR for each Eimeria species expressed as the number of oocysts/g droppings is presented in .
Inter-assay repeatability
The CV range of the qPCR was below 10% for all Eimeria spp. An exception was E. necatrix for which it ranged from 3.0% to 10.6%. The exact CV range for all the seven Eimeria spp. is given in .
Field validation
Broiler flocks and samples of droppings
An overview of the participating feed mills, farms, broiler flocks and results obtained by the qPCR using the pooled samples is given in . E. acervulina was detected in all 19 broiler flocks examined, E. maxima in 12 flocks, E. tenella in 13 flocks and E. mitis in one flock. E. praecox, E. necatrix and E. brunetti were not found in any of the flocks examined in this study.
Table 3. The participating feed mills, farms, broiler flocks monitored with the qPCR, sampling period, start, peak and end of oocyst shedding per Eimeria spp.
Shedding of oocysts started at variable time points. The start of E. acervulina oocyst excretion was observed between 13 and 31 days of age and frequently continued until the end of the grow-out period. On average, its excretion peaked at 28 days of age. Peak excretion ranged from 14 × 103 to 656 × 103 oocyst equivalents/g droppings.
The start of E. maxima oocyst excretion occurred between 13 and 32 days of age and continued until 30–46 days of age. On average, excretion peaked at 29 days of age. Peak excretion ranged from 0.2 × 103 to 16 × 103 oocyst equivalents/g droppings.
E. tenella oocyst excretion started between 16 and 34 days of age and continued until 37–46 days of age. Excretion peaked at approximately 34 days of age and ranged from 0.5 × 103 to 65 × 103 oocyst equivalents/g droppings.
E. mitis oocyst excretion was only observed in one flock where it started at 31 days of age and continued until the end of the grow-out period (41 days of age). Its excretion peaked at 36 days of age and ranged from 0.5 × 103 to 9.2 × 103 oocyst equivalents/g droppings.
Comparison between pooled and mixed samples
The correlation coefficients between the mean number of oocysts/g droppings and the oocyst equivalents/g droppings of the pooled samples and that of the corresponding mixed samples were 0.94 (95% CI: 0.92–0.95) and 0.87 (95% CI: 0.84–0.90) for the OPG-counting method and the qPCR, respectively ( and ).
Figure 2. Correlation between the mean number of oocysts/g dropping of the pooled samples and that of the corresponding mixed sample (n = 236) obtained by the OPG-counting method. Samples were obtained from 19 broiler flocks. One mixed sample consisted of 5 g dropping of each of five pooled samples (one pooled sample = 10 droppings) collected at the same time point.
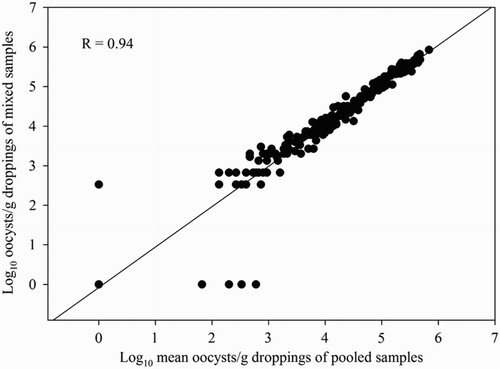
Figure 3. Correlation between the mean number of oocyst equivalents/g of the pooled samples and that of the corresponding mixed sample (n = 236) obtained by the multiplex Eimeria spp. qPCRs. Samples were obtained from 19 broiler flocks. One mixed sample consisted of 5 g dropping of each of five pooled samples (one pooled sample = 10 droppings) collected at the same time point.
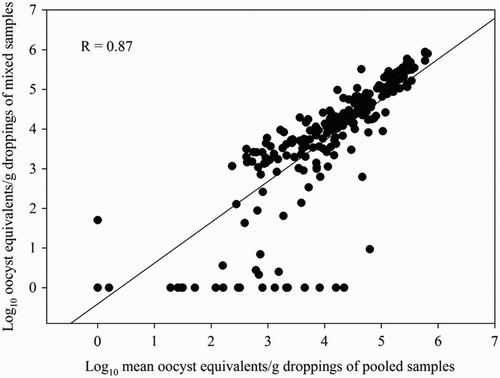
Only nine of the mixed samples were negative in both, the OPG-counting method and the qPCR, while the associated pooled samples yielded low numbers of oocysts.
Comparison between OPG-counts and the qPCR
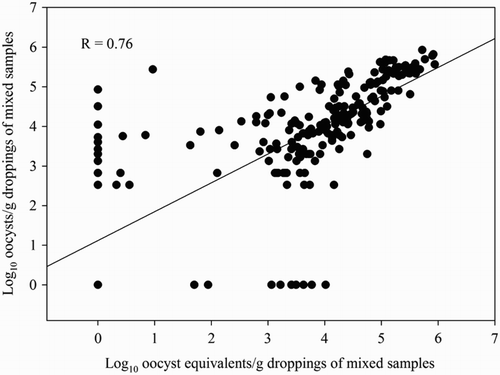
The Pearson’s correlation coefficients between both methods were 0.78 (95% CI: 0.76–0.80) and 0.76 (95% CI: 0.70 – 0.81) for both the pooled and mixed samples, respectively ( and ).
Figure 4. Correlation between the number of oocysts/g dropping of the pooled samples (n = 1180) obtained by the OPG-counting method and the number of oocyst equivalents/g dropping obtained by the multiplex Eimeria spp. qPCRs. Samples were obtained from 19 broiler flocks. One pooled sample = 10 droppings collected at the same time point.
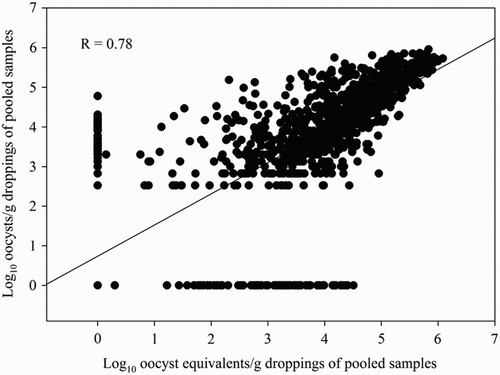
Figure 5. Correlation between the number of oocysts/g dropping of the mixed samples (n = 236) obtained by the OPG-counting method and the number of oocysts equivalents/g dropping obtained by the multiplex Eimeria spp. qPCRs. Samples were obtained from 19 broiler flocks. One mixed sample consisted of 5 g dropping of each of five pooled samples (one pooled sample = 10 droppings) collected at the same time point.
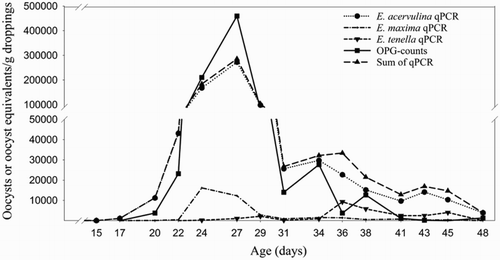
Despite differences in the absolute numbers of oocysts/g droppings of the OPG-counting method and the oocyst equivalents/g droppings of the qPCR, the graphs representing oocyst excretion/g droppings using either method during the grow-out period showed a good visual agreement (an example is given in ).
Figure 6. Example of Eimeria spp. oocyst shedding in one of the broiler flocks (farm C, flock 1) based on the mean of the OPG-counting method and the multiplex Eimeria spp. qPCRs results of pooled samples.
One thousand and eleven pooled samples showed positive results in the OPG-counting method or the qPCR or both. Of these, 934 were positive in the OPG-counting method and 952 were positive in the qPCR.
Seventy-seven pooled samples showed negative results in the OPG-counting method, of which 51 had a qPCR result of 3330 oocyst equivalents/g droppings or less. Fifty-nine pooled samples showed negative results in the qPCR, of which 40 samples yielded low amounts of oocysts in the OPG-counting method.
The number of mixed samples with positive results in the OPG-counting method, the qPCR or both amounted to 209. Nine were negative in the OPG-counting method, while the qPCR was positive. Seven of these samples contained less than 3330 oocyst equivalents per gram. Furthermore, 12 mixed samples were negative in the qPCR and positive in the OPG-counting method, of which seven contained less than 3330 OPG.
No explanation could be found for the remaining pooled or mixed samples that showed negative results in the qPCR but were positive in OPG-counting method, or the other way around when more than 3330 oocyst (equivalents) per gram were found.
Discussion
Monitoring of coccidiosis using lesion scores and oocyst excretion/g droppings is far from accurate for reasons mentioned in the introduction. Therefore, our aim was to further develop and validate a previously described Eimeria spp. qPCR for use on fresh dropping samples.
PCR inhibition occurred when we started to use an Eimeria spp. PCR directly on fresh droppings. In order to circumvent this, various techniques for DNA extraction were investigated (flotation, bleach, commercial kits, etc.) of which the NucleoSpin® Soilkit performed best. However, cross-reactivity between species was frequently found when using the PCR test according to Morgan et al. (Citation2009). Despite multiple adjustments, named cross-reactivity could not be prevented (non-published data). In 2010, Vrba and others published a PCR based on non-polymorphic SCARs, which we modified into three multiplex qPCRs for the detection of the seven chicken Eimeria spp. in fresh droppings. PCR efficiencies of the multiplex PCRs determined for individual species were ≥1.98, indicating a very high efficiency, which is comparable to the high efficiencies determined for the monoplex PCRs (Vrba et al., Citation2010). The analytical specificity (100%) of the primers and probes used were in agreement with the results of Vrba et al. (Citation2010). The analytical sensitivity of the qPCR was higher than that of the OPG-counting method (333 OPG droppings) for E. acervulina and E. tenella, but lower for E. maxima, E. mitis, E. brunetti, E. necatrix, and E. praecox. The lower analytical sensitivity of the latter may decrease the qPCR’s sensitivity during early or late infection; however, in the acute phase of a severe coccidiosis infection, a higher limit of detection should not be problematic. The inter-assay repeatability of the qPCR was considered good for all Eimeria spp. except E. necatrix, for which the quantitative results should be interpreted with caution.
The correlation coefficients between the mean number of oocysts/g droppings of the pooled samples and that of the corresponding mixed samples were 0.94 and 0.87 for the OPG-counting method and the qPCR, respectively, ( and ). These results indicate that the use of a mixed sample (composed of the five pooled samples) in coccidiosis monitoring programmes is a good alternative for the pooled samples.
Comparison of the OPG-counting method and the qPCR also showed a good, although slightly lower correlation of 0.78 and 0.76 for the pooled and mixed samples, respectively. The high correlation between both methods indicates that the qPCR can be used on fresh droppings for the monitoring of coccidiosis as a substitute for the traditional OPG-counting method.
The qPCR was successfully used for the monitoring of coccidiosis in 19 flocks housed on four different farms. The following Eimeria spp. were detected: E. acervulina (19 flocks), E. maxima (12 flocks), E. tenella (13 flocks), and E. mitis (one flock). Contrary to the traditional diagnostic methods (lesion scoring and OPG-counts), the qPCR enabled reliable species specific quantification of Eimeria parasites in fresh droppings. Thus, actual information on the start, height and duration of oocyst shedding can be easily obtained and may subsequently be used to study the effect of intervention strategies, e.g. anticoccidial programmes per Eimeria spp. An example was found in flock 2 of farm D, feed mill A (), where a delayed onset of the E. acervulina oocyst excretion (at 31 days of age) was found. This was explained by the switch to an anticoccidial product (decoquinate), which had not been used on this farm for a long time.
The Eimeria spp. qPCR described here showed a good analytical and diagnostic sensitivity as well as specificity. It also showed a low detection limit and good reproducibility (except for E. necatrix), and can therefore be used in a coccidiosis monitoring programme. However, in order to be able to interpret Eimeria spp. shedding patterns in the field, additional coccidiosis infection experiments are needed to study the correlation between Eimeria spp. oocyst shedding and flock performance.
Acknowledgments
We thank Constance Reugebrink for performing the qPCR assays, Dr Anouk Veldhuis for the statistical analysis and Dr J.H.H. van Eck for critically reading the manuscript.
Additional information
Funding
References
- Haug, A., Gjevrey, A., Thebo, P., Mattsson, J.G. & Kaldhusdal, M. (2008). Coccidial infections in commercial broilers: epidemiological aspects and comparison of Eimeria species identification by morphometric and polymerase chain reaction techniques. Avian Pathology, 37, 161–170. doi: 10.1080/03079450801915130
- Hodgson, J.N. (1970). Coccidiosis: oocyst counting technique for coccidiostat evaluation. Experimental Parasitology, 28, 99–102. doi: 10.1016/0014-4894(70)90073-1
- Johnson, J. & Reid, W.M. (1970). Anticoccidial drugs: lesion scoring techniques in battery and floor pen experiments with chickens. Experimental Parasitology, 28, 30–36. doi: 10.1016/0014-4894(70)90063-9
- McDougald, L.R. & Fitz-Coy, S.H. (2013). Coccidiosis. In D.E. Swayne, J.R. Glisson, L.R. McDougald, L.K. Nolan, D.L. Suarez & V.L. Nair (Eds.), Diseases of Poultry 13th edn (pp. 1148–1166). Ames, IA: Blackwell Publishing Professional.
- Morgan, J.A.T., Morris, G.M., Wlodek, B.M., Byrnes, R., Jenner, M., Constantinoiu, C.C., Anderson, G.R., Lew-Tabor, A.E., Molloy, J.B., Gasser, R.B. & Jorgensen, W.K. (2009). Real-time polymerase chain reaction (PCR) assays for the specific detection and quantification of seven Eimeria species that cause coccidiosis in chickens. Molecular and Cellular Probes, 23, 83–89. doi: 10.1016/j.mcp.2008.12.005
- OIE (World Organisation for Animal Health). (2017a). Chapter 3.6.3. Development and optimisation of nucleic acid detection assays. In Manual of Diagnostic Tests and Vaccines for Terrestrial Animals. Retrieved April 3, 2017, from http://www.oie.int/en/international-standard-setting/terrestrial-manual/access-online/
- OIE (World Organisation for Animal Health). (2017b). Chapter 3.6.5. Statistical approaches to validation. In Manual of Diagnostic Tests and Vaccines for Terrestrial Animals. Retrieved April 3, 2017, from http://www.oie.int/en/international-standard-setting/terrestrial-manual/access-online/
- Peek, H.W. & Landman, W.J.M. (2003). Resistance to anticoccidial drugs of Dutch avian Eimeria spp. field isolates originating from 1996, 1999 and 2001. Avian Pathology, 32, 391–401. doi: 10.1080/0307945031000121149
- Shirley, M.W., Smith, A.L. & Tomley, F.M. (2005). The biology of avian Eimeria with an emphasis on their control by vaccination. Advanced Parasitology, 60, 285–330. doi: 10.1016/S0065-308X(05)60005-X
- Stata Corporation. (2011). Stata Software Version 12. College Station, TX: Stata Corporation.
- Vrba, V., Blake, D.P. & Poplstein, M. (2010). Quantitative real-time PCR assays for detection and quantification of all seven Eimeria species that infect the chicken. Veterinary Parasitology, 174, 183–190. doi: 10.1016/j.vetpar.2010.09.006
