ABSTRACT
Vertebral osteomyelitis (VO) is a worldwide emerging disease that affects broilers. Recently, the isolation of Enterococcus faecalis in cases of the disease has been described. This study aimed at determining the genetic diversity and antimicrobial resistance profile of 12 E. faecalis strains isolated from broilers with VO. Strains were isolated from nine flocks from six farms in a high-density poultry production area in Southeast Brazil and were evaluated using multilocus sequence typing and phylogenetic analysis. Antimicrobial susceptibility tests and PCR were performed to detect antimicrobial resistance genes. E. faecalis isolates belonged to different sequence types (ST), six of which (ST49, ST100, ST116, ST202, ST249, and ST300) have been previously described. Strains ST708 and ST709 were newly identified in this study. Strain ST49 was most frequently isolated (50% of the flocks) from the analysed VO cases. No phylogenetic or phylogeographic relationship was found among the strains. The VO isolated E. faecalis strains showed highest resistance to aminoglycosides, mainly gentamicin (40%), but were highly susceptible to vancomycin (10%). Aminoglycoside resistance genes were detected in seven E. faecalis strains, and AAC6′-APH2″ genes were most frequently detected. The results showed that E. faecalis strains isolated from recently reported VO cases were highly diverse genetically. The diversity of genotypes in circulation in the analysed flocks, without apparent relationship among them, raises questions on aetiopathogenesis of the disease in broilers and evolutionary aspects of E. faecalis.
Introduction
Enterococcus spp. are ubiquitous, Gram-positive bacteria (Wages, Citation1998), with a worldwide distribution in avians and mammals, including humans (Tannock, Citation1995). In recent years, enterococci, particularly Enterococcus faecalis (Kola et al., Citation2010), have emerged as a major cause of nosocomial infections, leading to extra-intestinal infections in humans (Creti et al., Citation2004). Enterococci have an intrinsic resistance to many antibiotics and have also acquired new resistance genes, considering the vancomycin-resistant enterococci (VRE) (Cetinkaya et al., Citation2000; Willems & Bonten, Citation2007) that have become a major problem in nosocomial infections.
In poultry, enterococci may be associated with different diseases. Enterococcus spp. are frequently responsible for infection of the yolk sac in one-day-old chicks (Deeming, Citation2005). Enterococcus cecorum has been associated with vertebral osteomyelitis (VO) and/or arthritis in broilers worldwide (Devriese et al., Citation2002; Wood et al., Citation2002; Thayer et al., Citation2008; Herdt et al., Citation2009; Stalker et al., Citation2010; Makrai et al., Citation2011; Robbins et al., Citation2012; Aitchison et al., Citation2014), whereas Enterococcus durans was isolated from young chickens suffering from bacteraemia and encephalomalacia (Cardona et al., Citation1993; Abe et al., Citation2006). Moreover, Enterococcus hirae was reported to cause osteomyelitis (Kolbjørnsen et al., Citation2011) and endocarditis in broilers (Velkers et al., Citation2011) and focal cerebral necrosis (Devriese et al., Citation1991; Randall et al., Citation1993) and diarrhoea in chicks (Kondo et al., Citation1997). E. faecalis has been associated with systemic amyloidosis in laying hens (Landman et al., Citation1994) and broiler breeders (Steentjes et al., Citation2002) and with arthritis in domestic ducks (Bisgaard, Citation1981). Furthermore, a recent report of E. faecalis involved in cases of VO in broilers was described for the first time in Brazil (Braga et al., Citation2016), unlike most reports of this disease in broilers, which were mainly associated to E. cecorum. This observation raised the question of whether different or new E. faecalis clones exist or have emerged and might be associated with VO in Brazil. However, there is no information on genetic diversity of E. faecalis strains isolated from broilers with VO, so far.
As there is a lack of information on the characterization of E. faecalis involved with broilers with VO, molecular characterization and determination of antimicrobial susceptibility of these strains are essential. This would allow for comparison between human and bird bacterial strains. The zoonotic or non-zoonotic resistant bacteria in animals and their by-products represent a concern to animal and public health, because of the possibility of transmission of resistance genes to humans (Olsen et al., Citation2012). Therefore, this study aimed to provide information on the molecular characteristics and antimicrobial resistance profiles of E. faecalis isolated from broilers with VO.
Materials and methods
Samples
We analysed 12 previously described (Braga et al., Citation2016) E. faecalis strains that were isolated from natural cases of VO in broilers. The broilers belonged to nine different flocks and six municipalities from the largest poultry production area in Minas Gerais, Southeast Brazil. Briefly, after clinical evaluation, the broilers were necropsied and, for the confirmed VO cases, the caseous, necrotic material within the affected vertebral body of the fourth or free thoracic vertebra (Baumel, Citation1979; Hogg, Citation1984) was used for bacterial isolation and DNA extraction. Samples for bacterial isolation were collected aseptically from vertebral lesions using sterile swabs, whereas for DNA extraction and PCR, the necrotic, vertebral material was collected in sterile microtubes and frozen at −20°C. Histopathology was performed for all cases employing haematoxylin and eosin stain and Goodpasture’s staining solution, to identifying bacterial colonies associated with VO (Braga et al., Citation2016). The procedures in this study were approved by the Animal Experimentation Ethics Committee of Universidade Federal de Minas Gerais (Protocol 205/2011).
Bacterial isolation and identification
The bacteria were isolated and identified as previously described (Teixeira et al., Citation2007). Briefly, the vertebral lesion swabs were inoculated onto two sheep blood agar (BA) plates and one MacConkey agar plate. One BA plate and the MacConkey agar plate were incubated under aerobic conditions at 37°C for 24–72 h. The other BA plate was incubated under microaerophilic conditions at the same temperature and time. The isolated colonies were Gram stained and submitted to catalase and oxidase tests. Bacterial isolates were subjected to automatic bacterial identification by VITEK 2 system (bioMérieux, Inc., Hazelwood, MO, USA), according to the manufacturer’s recommendations.
DNA extraction and PCR of E. faecalis from lesions and cultures
To confirm the presence of E. faecalis in the lesions, as detailed by Braga et al. (Citation2016), total DNA was extracted directly from the caseonecrotic material obtained from vertebral lesions, using a previously described method (Vogelstein & Gillespie, Citation1979; Boom et al., Citation1990). Briefly, tissue samples were ground in a mortar and pestle in three volumes of 6 M sodium iodide (Synth, São Paulo, Brazil), and the total DNA was recovered on silicon dioxide microspheres (Sigma-Aldrich, St. Louis, MO, USA).
Additionally, E. faecalis DNA was extracted directly from the bacterial colonies isolated from VO lesions to perform multilocus sequence typing (MLST), and aminoglycoside resistance gene (ARG) PCR. E. faecalis reference colonies (CCCD-E006, Cefar Diagnostica Ltd., São Paulo, Brazil) were used as control for PCR. DNA extraction was performed by boiling as described previously (Marques & Suzart, Citation2004), with some modifications. Briefly, multiple E. faecalis colonies were selected and transferred into a sterile microtube containing 300 µl ultrapure water and homogenised by vortexing (10 s). The microtubes were incubated at 100°C in a dry bath (30 min) and centrifuged at 14,000×g (2 min), and the supernatant was transferred to a new microtube. The quantity and purity of the DNA extracted were assessed using Nanodrop spectrophotometer (Thermo Scientific, Wilmington, DE, USA), and the DNA was stored at −20°C.
The extracted DNA was subjected to PCR using primers specific for E. faecalis () to amplify a region of sodA gene using previously described amplification protocols (Jackson et al., Citation2004; Braga et al., Citation2016).
Table 1. Primer sequences used for the diagnosis of E. faecalis and detection of ARGs in the isolates from broilers with VO, using PCR.
Multilocus sequence typing
The genetic relationships among E. faecalis strains were determined by MLST, as previously described (Ruiz-Garbajosa et al., Citation2006). The oligonucleotide sequences and PCR conditions mentioned on the E. faecalis MLST homepage (http://pubmlst.org/efaecalis/) were used. PCR was performed on 25 µl final volume using 250 ng DNA template, 20 pmol of each primer, and 12.5 µl PCR Master Mix (Promega, Madison, WI, USA). The following PCR conditions were used: 94°C for 5 min; 30 cycles of 94°C for 30 s, 52°C for 30 s, and 72°C for 1 min; and a final extension at 72°C for 7 min. Sequence types (ST) were defined based on the housekeeping gene sequences in the E. faecalis MLST database (http://pubmlst.org/efaecalis). The eBURST V3 program (http://eburst.mlst.net) was used to identify clonal complexes and to construct a population snapshot of all published STs (http://eburst.mlst.net/v3/mlst_datasets/). New alleles were deposited within the E. faecalis MLST database.
Antimicrobial susceptibility profile
E. faecalis isolated from VO lesions were submitted to antimicrobial susceptibility tests, as described by the Clinical and Laboratory Standards Institute (CLSI, Citation2013). The antimicrobial classes and their concentrations per disk (Cefar Diagnostica Ltda., São Paulo, Brazil) were as follows: aminoglycosides (neomycin, 30 μg; gentamicin, 10 μg; gentamicin high-level aminoglycoside resistance (HLAR), 120 μg; and streptomycin HLAR, 300 μg); beta-lactams (amoxicillin, 10 μg; ampicillin, 10 μg; ceftiofur, 30 μg; and penicillin/novobiocin, 40 μg); polypeptides (bacitracin, 10 IU); and glycopeptides (vancomycin, 30 μg). The inhibition zone diameters were measured and the values obtained were interpreted according to the CLSI (Citation2013) guidelines, as resistant, intermediate, or sensitive.
Aminoglycoside resistance genes
Multiplex PCR was conducted on bacterial colonies DNA for detection of six ARGs in enterococci (), as previously described (Vakulenko et al., Citation2003), with some modifications. PCR was performed using PCR Master Mix (Promega, Madison, WI, USA) and 250 ng template DNA, on a final volume of 50 µL. The PCR products were subjected to 1.5% agarose gel electrophoresis, stained with ethidium bromide (0.5 µg/ml for 40 min), and visualized using a UV transilluminator. The LowRanger 100 bp DNA Ladder (Norgen Biotek Corp., Thorold, ON, Canada) was used to estimate the molecular size of the PCR products.
The ARG amplicons were purified and sequenced using the Big-Dye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, Foster City, CA, USA) on an ABI 3130 Genetic Analyzer (Applied Biosystems). The obtained sequences were analysed using BioEdit Sequence Alignment Editor to acquire the consensus sequences, which were used for comparisons against GenBank database (NCBI) sequences, to confirm the detection of aminoglycoside genes in E. faecalis strains.
Results
E. faecalis isolation from VO
Bacterial isolation and identification tests allowed us to confirm the presence of E. faecalis in all VO cases analysed in this study. In addition to this bacterium, Escherichia coli or Staphylococcus aureus were isolated from other VO cases, which were not included in this study (Braga et al., Citation2016).
Gross and histopathology
E. faecalis strains were isolated from the T4 vertebra of broilers with osteomyelitis. Clinicopathological findings of these cases have been detailed by Braga et al. (Citation2016). Briefly, the affected broilers presented impaired mobility correlated to the spinal cord compression caused by swelling of the vertebral body due to infectious osteomyelitis. Sagittal section of the T4 region revealed whitish to yellowish, friable, caseonecrotic material replacing the normal vertebral body and compression of the spinal cord. On the histopathology, there were necrosis and inflammation of the vertebral body often associated with intralesional bacterial colonies.
MLST
The seven housekeeping genes of the 12 strains were successfully amplified and sequenced. MLST analysis revealed high genetic diversity of E. faecalis. Eight different STs were detected among the strains isolated from broilers with VO (). Five E. faecalis strains isolated from three flocks (1, 4, and 8) from different municipalities were ST49 (), which is the founder of a clonal complex that also includes ST203 and ST309 (). The other seven E. faecalis strains belonged to seven distinct STs, five of which have been previously described in the E. faecalis MLST database, namely ST100, ST116, ST202, ST249, and ST300. The other two STs were not detected previously, and were described for the first time in this study. They were deposited in the E. faecalis online database under the numbers ST708 and ST709. These E. faecalis isolates were the only singletons among the strains analysed ().
Figure 1. Geographical distribution of E. faecalis isolated from broilers with VO from Southeast Brazil, in 2012. A, B, C, D, E, and F represent the different municipalities included in this study, which are linked to their respective boxes with details of the strains isolated from the place (strain ID, number of the flock, and sequence type number). Distance between the farms was: A–F (130 km); F–B (47 km); B–E (45 km); E–C (42 km); C–D (54 km); and D–A (161 km), comprising a total area of 10,434 km2.
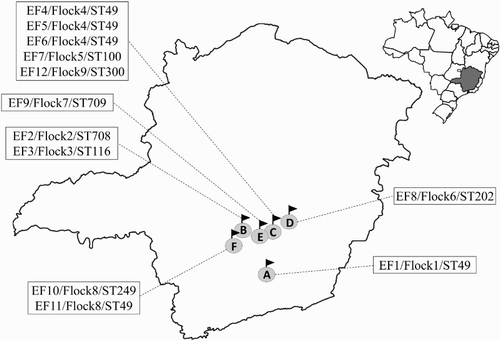
Figure 2. Population snapshot of E. faecalis STs available in the MLST database, including strains isolated from broilers with VO from Southeast Brazil, in 2012. Each ST is represented as a dot with the ST number. Clusters of linked STs correspond to clonal complexes. Black lines connect single locus variants. Primary founders are positioned centrally in the cluster. Gray arrows indicate clusters with STs available in the E. faecalis database that were also identified among the isolates described in this study. STs pointed by black arrows are newly described in this study.
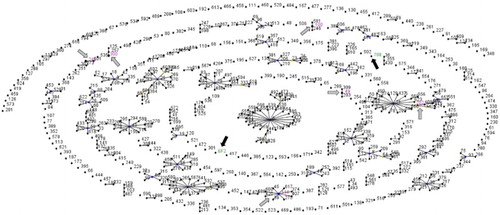
Table 2. ST, antimicrobial susceptibility profile and ARGs detected by PCR of E. faecalis isolated from VO in broilers from Minas Gerais, Brazil (2012).
These results showed that different genotypes of E. faecalis caused the disease in distinct flocks and municipalities in the same year ().
Antimicrobial susceptibility test
Antimicrobial susceptibility profile of 10 E. faecalis strains is shown in . Seven E. faecalis isolates were resistant to at least one aminoglycoside drug. The highest resistance levels were found against gentamicin and neomycin, in four and three strains, respectively. Five isolates had a high-level of aminoglycoside resistance, characterized by streptomycin HLAR in three and gentamicin HLAR in two E. faecalis strains. Vancomycin resistance was seen in only one E. faecalis strain. All E. faecalis isolates from vertebral lesions were sensitive to ampicillin, amoxicillin, and penicillin/novobiocin.
The highest multiple antimicrobial resistance index was 30% (3/10), observed in one E. faecalis strain that was resistant to neomycin, streptomycin HLAR, and ceftiofur. Four strains (EF2, EF4, EF6, and EF8) had multiple antimicrobial resistance index of 20% (2/10), characterized by resistance to different combinations of two aminoglycoside antimicrobials and represented as gentamicin/gentamicin aminoglycoside-resistant (EF4 and EF8) or neomycin/streptomycin-resistant (EF2 and EF6) strains. Three E. faecalis strains (EF5, EF7, and EF11) were resistant to only one antimicrobial: gentamicin (EF5 and EF11) or vancomycin (EF7), whereas two strains (EF3 and EF9) were susceptible to all the tested antimicrobials.
ARG profiles of E. faecalis strains
ARGs were detected in the seven E. faecalis strains (). The 6′-aminoglycoside acetyltransferase and 2″-aminoglycoside phosphotransferase (AAC6′-APH2″) genes were present in EF5, EF6, EF10, and EF11, whereas in EF4, EF8, and EF12 only the aminoglycoside 3′-phosphotransferase type IIIa gene was present. The ARG PCR products were confirmed by sequencing and alignment using BLASTN.
Discussion
These are the first data regarding molecular typing of E. faecalis isolated from VO in broilers. The results showed high genetic diversity among the isolated E. faecalis, as evidenced by the different STs obtained (8) proportional to the strains analysed (12). Two different situations were noted: the detection of the same ST (ST49) in three different VO cases in the same flock (Flock 4, ) and the detection of different STs (ST49 and ST249) in two different disease cases in the same flock (Flock 8, ). Although no unique E. faecalis STs were isolated from VO cases in different flocks, ST49 was most frequently detected.
E. faecalis ST49 was previously detected in broiler breeder flocks, in two apparently healthy broiler breeders (Gregersen et al., Citation2010). E. faecalis described in this study were isolated from VO cases that had Gram-positive cocci associated with the lesions, confirmed by histopathology special stains (Braga et al., Citation2016). Recently, it was observed that ST49 are more frequently detected in hospitalized human patients than in non-hospitalized human patients (Tedim et al., Citation2015). Considering the importance of E. faecalis in humans, pursuing further studies to assess the role of broilers in its epidemiology remains relevant.
The association between E. faecalis STs and lesion types still seems to be conflicting. Gregersen et al. (Citation2010) observed no correlation between E. faecalis STs and lesion types in broiler breeders. On the other hand, Petersen et al. (Citation2009) analysed E. faecalis strains associated with arthritis and amyloid arthropathy in laying hens from five different countries and reported that 71.4% (15/21) of these strains were ST82. This ST was not among the E. faecalis strains isolated in our study nor, to the author’s knowledge, among other Brazilian E. faecalis strains.
E. faecalis ST249 was identified in one case of VO analysed. This ST was also detected in two apparently healthy and three sick broilers (Gregersen et al., Citation2010). The other STs (ST100, ST116, ST202, ST249, and ST300) have already been described for E. faecalis isolated elsewhere. ST116, ST202, and ST300 were isolated from retail chicken meat in Korea (Choi & Woo, Citation2013), although there is no report of these STs related to disease in the chicken. Moreover, E. faecalis ST116 was isolated from a hospitalized human patient in Cuba (Quiñones et al., Citation2009), whereas ST100 was identified from swine in Denmark (Shankar et al., Citation2006). To the author’s knowledge, there is no description of ST100 infection in chickens.
Our results indicate that in high-density poultry production areas, different broiler flocks of same or different farms may be infected contemporaneously with genetically different E. faecalis strains. They also point out the importance of VO diagnosis in the region, to determine the tendency of its occurrence over the years, and of molecular profiling of E. faecalis strains. MLST performed in this study analysed the sequences of seven housekeeping genes for E. faecalis. MLST still is one of the typing methods of choice for performing bacterial population genetics studies (Maiden et al., Citation1998), which can provide valuable information on the evolution and diversification of these species (Belkum et al., Citation2007). However, this tool may not provide the same level of resolution as the whole genome data, since its analysis is based on limited information obtained from a fraction of the genome (SenGupta et al., Citation2014). This may limit the identification of precise epidemiological relationships (Köser et al., Citation2012), mainly those occurring in short periods of time (SenGupta et al., Citation2014).
E. faecalis is part of the intestinal microbiota of chickens, just like E. cecorum, which is often described to be associated with the disease. Although E. cecorum has been studied as a cause of VO in broilers, the factors leading to increased incidence of infections caused by this bacterium are not fully understood. According to Boerlin et al. (Citation2012), the main hypotheses to be considered are changes occurring in the host and environmental factors or the emergence of individual clones with increased pathogenicity. Recently, Borst et al. (Citation2016) studied the pathogenesis of VO caused by E. cecorum in broiler and their results suggested that some events such as intestinal colonization, bacteraemia, and osteochondrosis dissecans of the free thoracic vertebra in early life are crucial to the pathogenesis of the disease. To the author’s knowledge, no information on pathogenesis study or molecular typing of E. faecalis involved in VO is available, highlighting the need for studies that could help to understand the aetiopathogenesis of the disease in broilers.
HLAR was detected in half of the E. faecalis isolates. This trait was detected in ST49 (2/5) and ST100 (1/5), as well as in ST708 (1/5) and ST709 (1/5) described in this study. High-level gentamicin resistance was also observed in 10.9% (11/101) of the food-borne E. faecalis isolated from retail chicken meat in Korea (Choi & Woo, Citation2013).
High-level gentamicin resistance is associated with bifunctional AAC6′-APH2″ aminoglycoside-modifying enzymes, which reduce the effect of aminoglycosides (Udo et al., Citation2004). Streptomycin is an exception and is modified by 6-nucleotidyltransferase (Chow, Citation2000). In our study, AAC6′-APH2″ genes were detected in four of seven E. faecalis strains considered to be aminoglycoside-resistant based on the antimicrobial susceptibility tests, whereas three strains presented the aminoglycoside 3′-phosphotransferase type IIIa gene ().
Vancomycin resistance was detected only in EF8 E. faecalis strain (ST202). In enterococci, vancomycin resistance is encoded by different genes, such as vanA, which was detected in E. faecalis isolated from meat or birds associated with poultry production in Asia and New Zealand (Agersø et al., Citation2008). Interestingly, VRE strains isolated from humans, chicken, and swine in Malaysia belonged to six different STs, but none of them was ST202. In addition, the authors of this study highlighted the infrequent detection of a human VRE clone in these broilers.
Antimicrobial resistance genes in enterococci are a danger to animal and human health, especially those located on mobile genetic elements, because these bacteria could transfer these genes to other potentially pathogenic bacteria in the chicken intestine and also to zoonotic bacteria. Moreover, these enterococci might be transferred to humans, causing disease and/or further spreading the resistance genes to the gastrointestinal bacteria (Cauwerts et al., Citation2007). It has already been demonstrated that E. faecalis of human and poultry origin share virulence genes, supporting the zoonotic potential of E. faecalis (Olsen et al., Citation2012).
E. faecalis can acquire antimicrobial resistance through transfer of plasmids and transposons, chromosomal exchange, or mutations (Coque, Citation2008). Moreover, it can also act as a source of antimicrobial resistance genes for poultry intestinal pathogens (Donelli et al., Citation2004). This highlights its role in the generation of antimicrobial resistance and its potential implication in bird and human health.
Our results showed a high genetic diversity among these strains, with two new STs (ST708 and ST709) described for the first time in this study, and a lack of epidemiological relationship among these isolates. As VO in broilers has been reported to be most frequently associated with E. cecorum and the involvement of E. faecalis in this disease has been reported only recently, experimental infection studies of these E. faecalis strains are needed, which, in addition, may represent emergent clones. Additional studies on E. faecalis could help elucidate the questions, such as the aetiopathogenesis of VO in broilers, evolutionary aspects of E. faecalis, and role of antibiotic use in birds and humans and their impact on veterinary poultry production and human health.
Acknowledgements
We would like to thank Pró-Reitoria de Pesquisa of Universidade Federal de Minas Gerais and Dr Liliane Denize Miranda Menezes (Instituto Mineiro de Agropecuária) for helping with bacterial identification.
Disclosure statement
No potential conflict of interest was reported by the authors.
ORCID
Nelson Rodrigo da Silva Martins http://orcid.org/0000-0001-8925-2228
Roselene Ecco http://orcid.org/0000-0002-8052-5389
Additional information
Funding
References
- Abe, Y., Nakamura, K., Yamada, M. & Yamamoto, Y. (2006). Encephalomalacia with Enterococcus durans infection in the brain stem and cerebral hemisphere in chicks in Japan. Avian Diseases, 50, 139–141.
- Agersø, Y., Lester, C.H., Porsbo, L.J., Ørsted, I., Emborg, H.D., Olsen, K.E.P., Jensen, L.B., Heuer, O.E., Frimodt-Møller, N., Aarestrup, F.M. & Hammerum, A.M. (2008). Vancomycin-resistant Enterococcus faecalis isolates from a Danish patient and two healthy human volunteers are possibly related to isolates from imported turkey meat. Journal of Antimicrobial Chemotherapy, 62, 844–845.
- Aitchison, H., Poolman, P., Coetzer, M., Griffiths, C., Jacobs, J., Meyer, M. & Bisschop, S. (2014). Enterococcal-related vertebral osteoarthritis in South African broiler breeders: a case report. Journal of the South African Veterinary Association, 85, 1077.
- Baumel, J.J. (1979). Osteología. In J.J. Baumel, A.S. King, A.M. Lucas, J.E. Breazile & H.E. Evans (Eds.). Nomina Anatomica Avium (pp. 53–121). London: Academic Press.
- Belkum, A.V., Tassios, P.T., Dijkshoorn, L., Haeggman, S., Cookson, B., Fry, N.K., Fussing, V., Green, J., Feil, E., Gerner-Smidt, P., Brisse, S. & Struelens, M. (2007). Guidelines for the validation and application of typing methods for use in bacterial epidemiology. Clinical Microbiology and Infectious Diseases, 13, 1–46.
- Bisgaard, M. (1981). Arthritis in ducks: aetiology and public health aspects. Avian Pathology, 10, 11–21.
- Boerlin, P., Nicholson, V., Brash, M., Slavic, D., Boyen, F., Sanei, B. & Butaye, P. (2012). Diversity of Enterococcus cecorum from chickens. Veterinary Microbiology, 157, 405–411.
- Boom, R., Sol, C.J., Salimans, M.M., Jansen, C.L., Dillen, P.M.W.-V. & Noordaa, J.V.D. (1990). Rapid and simple method for purification of nucleic acids. Journal of Clinical Microbiology, 28, 495–503.
- Borst, L.B., Suyemoto, M.M., Sarsour, A.H., Harris, M.C., Martin, M.P., Strickland, J.D., Oviedo, E.O. & Barnes, H.J. (2017). Pathogenesis of enterococcal spondylitis caused by Enterococcus cecorum in broiler chickens. Veterinary Pathology, 54, 61–73.
- Braga, J.F.V., Silva, C.C., Teixeira, M.P., Martins, N.R. & Ecco, R. (2016). Vertebral osteomyelitis associated with single and mixed bacterial infection in broilers. Avian Pathology, 45, 640–648.
- Cardona, C.J., Bickford, A.A., Charlton, B.R. & Cooper, G.L. (1993). Enterococcus durans infection in young chickens associated with bacteremia and encephalomalacia. Avian Diseases, 37, 234–239.
- Cauwerts, K., Decostere, A., De Graef, E.M., Haesebrouck, F. & Pasmans, F. (2007). High prevalence of tetracycline resistance in Enterococcus isolates from broilers carrying the erm(B) gene. Avian Pathology, 36, 395–399.
- Cetinkaya, Y., Falk, P. & Mayhall, C.G. (2000). Vancomycin-resistant enterococci. Clinical Microbiology Reviews, 13, 686–707.
- Choi, J.M. & Woo, G.J. (2013). Molecular characterization of high-level gentamicin-resistant Enterococcus faecalis from chicken meat in Korea. International Journal of Food Microbiology, 165, 1–6.
- Chow, J.W. (2000). Aminoglycoside resistance in enterococci. Clinical Infectious Diseases, 31, 586–589.
- Clinical and Laboratory Standards Institute. (2013). VET01-A4, performance standards for antimicrobial disk and dilution susceptibility tests for bacteria isolated from animal; Approved Standard-Fourth Edition. [Internet].
- Coque, T.M. (2008). Evolutionary biology of pathogenic enterococci. In F. Baquero, C. Nombela, G.H. Cassell & J.A. Guitierrez (Eds.). Evolutionary biology of bacterial and fungal pathogens (pp. 501–521). Washington, DC: ASM Press.
- Creti, R., Imperi, M., Bertuccini, L., Fabretti, F., Orefici, G., Di Rosa, R. & Baldassarri, L. (2004). Survey for virulence determinants among Enterococcus faecalis isolated from different sources. Journal of Medical Microbiology, 53, 13–20.
- Deeming, D.C. (2005). Yolk sac, body dimensions and hatchling quality of ducklings, chicks and poults. British Poultry Science, 46, 560–564.
- Devriese, L.A., Cauwerts, K., Hermans, K. & Wood, A.M. (2002). Enterococcus cecorum septicemia as a cause of bone and joint lesions resulting in lameness in broiler chickens. Vlaams Diergeneeskundig Tijdschrift, 71, 219–221.
- Devriese, L.A., Hommez, J., Wijfels, R. & Haesebrouck, F. (1991). Composition of the enterococcal and streptococcal intestinal flora of poultry. Journal of Applied Bacteriology, 71, 46–50.
- Donelli, G., Paoletti, C., Baldassarri, L., Guaglianone, E., Di Rosa, R., Magi, G., Spinaci, C. & Facinelli, B. (2004). Sex pheromone response, clumping, and slime production in enterococcal strains isolated from occluded biliary stents. Journal of Clinical Microbiology, 42, 3419–3427.
- Gregersen, R.H., Petersen, A., Christensen, H. & Bisgaard, M. (2010). Multilocus sequence typing of Enterococcus faecalis isolates demonstrating different lesion types in broiler breeders. Avian Pathology, 39, 435–440.
- Herdt, P., Defoort, P., Van Steelant, J., Swam, H., Tanghe, L., Van Goethem, S. & Vanrobaeys, M. (2009). Enterococcus cecorum osteomyelitis and arthritis in broiler chickens. Vlaams Diergeneeskundig Tijdschrift, 78, 44–48.
- Hogg, D.A. (1984). The distribution of pneumatisation in the skeleton of the adult domestic fowl. Journal of Anatomy, 138, 617–629.
- Jackson, C.R., Fedorka-Cray, P.J. & Barrett, J.B. (2004). Use of a genus- and species-specific multiplex PCR for identification of enterococci. Journal of Clinical Microbiology, 42, 3558–3565.
- Kola, A., Schwab, F., Barwolff, S., Eckmanns, T., Weist, K., Dinger, E., Klare, I., Witte, W., Ruden, H. & Gastmeier, P. (2010). Is there an association between nosocomial infection rates and bacterial cross transmissions? Critical Care Medicine, 38, 46–50.
- Kolbjørnsen, Ø., David, B. & Gilhuus, M. (2011). Bacterial osteomyelitis in a 3-week-old broiler chicken associated with Enterococcus hirae. Veterinary Pathology, 48, 1134–1137.
- Kondo, H., Abe, N., Tsukuda, K. & Wada, Y. (1997). Adherence of Enterococcus hirae to the duodenal epithelium of chicks with diarrhoea. Avian Pathology, 26, 189–194.
- Köser, C.U., Ellington, M.J., Cartwright, E.J., Gillespie, S.H., Brown, N.M., Farrington, M., Holden, M.T., Dougan, G., Bentley, S.D., Parkhill, J. & Peacock, S.J. (2012). Routine use of microbial whole genome sequencing in diagnostic and public health microbiology. PLoS Pathogens, 8, e1002824.
- Landman, W.J., Gruys, E. & Dwars, R.M. (1994). A syndrome associated with growth depression and amyloid arthropathy in layers: a preliminary report. Avian Pathology, 23, 461–470.
- Maiden, M.C.J., Bygraves, J.A., Feil, E., Morelli, G., Russell, J.E., Urwin, R., Zhang, Q., Zhou, J., Zurth, K., Caugant, D.A., Feavers, I.M., Achtman, M. & Spratt, B.G. (1998). Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proceedings of the National Academy of Sciences of the United States of America, 95, 3140–3145.
- Makrai, L., Nemes, C., Simon, A., Ivanics, E., Dudás, Z., Fodor, L. & Glávits, R. (2011). Association of Enterococcus cecorum with vertebral osteomyelitis and spondylolisthesis in broiler parent chicks. Acta Veterinaria Hungarica, 59, 11–21.
- Marques, E.B. & Suzart, S. (2004). Occurrence of virulence-associated genes in clinical Enterococcus faecalis strains isolated in Londrina, Brazil. Journal of Medical Microbiology, 53, 1069–1073.
- Olsen, R.H., Schønheyder, H.C., Christensen, H. & Bisgaard, M. (2012). Enterococcus faecalis of human and poultry origin share virulence genes supporting the zoonotic potential of E. faecalis. Zoonoses and Public Health, 59, 256–263.
- Petersen, A., Christensen, H., Philipp, H.-C. & Bisgaard, M. (2009). Clonality of Enterococcus faecalis associated with amyloid arthropathy in chickens evaluated by multilocus sequence typing (MLST). Veterinary Microbiology, 134, 392–395.
- Quiñones, D., Kobayashi, N. & Nagashima, S. (2009). Molecular epidemiologic analysis of Enterococcus faecalis isolates in Cuba by multilocus sequence typing. Microbial Drug Resistance, 15, 287–293.
- Randall, C.J., Wood, A.M. & MacKenzie, G. (1993). Encephalomalacia in first-week chicks. Veterinary Record, 132, 419–419.
- Robbins, K.M., Suyemoto, M.M., Lyman, R.L., Martin, M.P., Barnes, H.J. & Borst, L.B. (2012). An outbreak and source investigation of enterococcal spondylitis in broilers caused by Enterococcus cecorum. Avian Diseases, 56, 768–773.
- Ruiz-Garbajosa, P., Bonten, M.J., Robinson, D.A., Top, J., Nallapareddy, S.R., Torres, C., Coque, T.M., Cantón, R., Baquero, F., Murray, B.E., del Campo, R. & Willems, R.J.L. (2006). Multilocus sequence typing scheme for Enterococcus faecalis reveals hospital-adapted genetic complexes in a background of high rates of recombination. Journal of Clinical Microbiology, 44, 2220–2228.
- SenGupta, D.J., Cummings, L.A., Hoogestraat, D.R., Butler-Wu, S.M., Shendure, J., Cookson, B.T. & Salipante, S.J. (2014). Whole-genome sequencing for high-resolution investigation of methicillin-resistant Staphylococcus aureus epidemiology and genome plasticity. Journal of Clinical Microbiology, 52, 2787–2796.
- Shankar, N., Baghdayan, A.S., Willems, R., Hammerum, A.M. & Jensen, L.B. (2006). Presence of pathogenicity island genes in Enterococcus faecalis isolates from pigs in Denmark. Journal of Clinical Microbiology, 44, 4200–4203.
- Stalker, M.J., Brash, M.L., Weisz, A., Ouckama, R.M. & Slavic, D. (2010). Arthritis and osteomyelitis associated with Enterococcus cecorum infection in broiler and broiler breeder chickens in Ontario, Canada. Journal of Veterinary Diagnostic Investigation, 22, 643–645.
- Steentjes, A., Veldman, K.T., Mevius, D.J. & Landman, W.J. (2002). Molecular epidemiology of unilateral amyloid arthropathy in broiler breeders associated with Enterococcus faecalis. Avian Pathology, 31, 31–39.
- Tannock, G.W. (1995). Normal microflora: an introduction to microbes inhabiting the human body. London: Chapman and Hall.
- Tedim, A.P., Ruiz-Garbajosa, P., Corander, J., Rodríguez, C.M., Cantón, R., Willems, R.J., Baquero, F. & Coque, T.M. (2015). Population biology of intestinal Enterococcus isolates from hospitalized and nonhospitalized individuals in different age groups. Applied and Environmental Microbiology, 81, 1820–1831.
- Teixeira, L., Carvalho, M. & Facklan, R. (2007). Enterococcus. In P. Murray, E. Baron, M. Landry, J. Jorgensen & M. Pfaller (Eds.). Manual of Clinical Microbiology 9th edn (pp. 430–442). Washington, DC: ASM Press.
- Thayer, S.G., Waltman, W.D. & Wages, D.P. (2008). Streptococcus and Enterococcus. In Y.M. Saif, A.M. Fadly, J.R. Glisson, L.R. McDougald, L.K. Nolan & D.E. Swayne (Eds.). Diseases of Poultry 12th edn (pp. 900–908). Ames, IA: Blackwell Publishing.
- Udo, E.E., Al-Sweih, N., John, P., Jacob, L.E. & Mohanakrishnan, S. (2004). Characterization of high-level aminoglycoside-resistant enterococci in Kuwait hospitals. Microbial Drug Resistance, 10, 139–145.
- Vakulenko, S.B., Donabedian, S.M., Voskresenskiy, A.M., Zervos, M.J., Lerner, S.A. & Chow, J.W. (2003). Multiplex PCR for detection of aminoglycoside resistance genes in enterococci. Antimicrobial Agents and Chemotherapy, 47, 1423–1426.
- Velkers, F.C., Graaf-Bloois, L.V., Wagenaar, J.A., Westendorp, S.T., van Bergen, M.A., Dwars, R.M. & Landman, W.J. (2011). Enterococcus hirae-associated endocarditis outbreaks in broiler flocks: clinical and pathological characteristics and molecular epidemiology. Veterinary Quarterly, 31, 3–17.
- Vogelstein, B. & Gillespie, D. (1979). Preparative and analytical purification of DNA from agarose. Proceedings of the National Academy of Sciences of the United States of America, 76, 615–619.
- Wages, D.P. (1998). Streptococcosis. In D.E. Swayne, J.R. Glisson, M.W. Jackwood, J.E. Person, & W.M. Reed, (Eds.). Isolation and Identification of Avian Pathogens 4th edn (pp. 58–60). Kennett Square, PA: American Association of Avian Pathologists.
- Willems, R.J. & Bonten, M.J. (2007). Glycopeptide-resistant enterococci: deciphering virulence, resistance and epidemicity. Current Opinion in Infectious Diseases, 20, 384–390.
- Wood, A.M., Mackenzie, G., Mcgiliveray, N.C., Brown, L., Devriese, L.A. & Baele, M. (2002). Isolation of Enterococcus cecorum from bone lesions in broiler chickens. Veterinary Record, 150, 27.
