ABSTRACT
We performed pathological and molecular virological investigation of three outbreaks of highly pathogenic avian influenza (HPAI) in a quail farm and two duck farms of Mymensingh and Netrokona districts of Bangladesh in 2011. HPAI viruses of subtype H5N1 were detected from all three outbreaks and phylogenetic analysis of HA gene sequence placed the viruses into clade 2.3.2.1. The outbreak in the quail farm was characterized by acute death with 100% mortality within two days. Marked haemorrhages and congestion with necrotic and inflammatory lesions in the respiratory tract, liver, pancreas and kidneys were the major gross and histopathological lesions. In the case of ducks, nervous signs were the remarkable clinical manifestations and the mortality was around 10%. No significant gross lesions were observed at necropsy. Non-purulent encephalitis with gliosis and neuronal degeneration was observed on histopathological examination. By immunohistochemistry, viral antigen could be detected in different organs of both quails and ducks. This study records varying clinical and pathological manifestations of HPAI in ducks and quails following natural infection with the same strain of the virus.
RESEARCH HIGHLIGHTS
HPAIV of clade 2.3.2.1 was detected from clinical outbreaks in quails and ducks
Sudden death with severe haemorrhages in various organs was found in quails
Pronounced nervous signs with non-purulent encephalitis were observed in ducks
Viral antigen could be localized in different organs by immunohistochemistry
Introduction
Avian influenza (AI) is an important viral disease of poultry caused by influenza A viruses of the family Orthomyxoviridae having a single-stranded negative sense segmented RNA genome. Type A influenza viruses can be classified into several subtypes based on their major surface proteins, haemagglutinin (HA) and neuraminidase (NA). So far, 16 HA subtypes (H1–H16) and nine NA subtypes (N1–N9) have been detected in birds (Webster et al., Citation1992; Fouchier et al., Citation2005). Influenza viruses of the H5 and H7 subtypes often emerge as virulent viruses and are commonly referred to as highly pathogenic avian influenza (HPAI) (Guan et al., Citation2004). Since its first detection in 1996, the present panzootic HPAI virus (HPAIV) of subtype H5N1 has spread to over 60 countries in Asia, Europe and Africa infecting wild birds and domestic poultry with sporadic zoonotic transmission to humans (Guan et al., Citation2004). The H5 avian influenza viruses are under continuous evolution, and molecular phylogenetic analysis revealed 10 distinct HA clades (0–9) and several sub-clades (Cattoli et al., Citation2011; WHO, Citation2014).
Bangladesh experienced the first outbreak of H5N1 HPAI in 2007. Since then the virus has become entrenched in poultry farms of Bangladesh and more than 550 outbreaks of avian influenza were reported to the World Organization for Animal Health (OIE, Citation2017). The majority of the avian influenza outbreaks were limited to chickens only with occasional spill-over to feral crows; however, a few outbreaks in other bird species such as ducks, quails and migratory birds were also recorded (Islam et al., Citation2012; Khan et al., Citation2014; Parvin et al., Citation2014; Sarker et al., Citation2017; Haider et al., Citation2017). Several studies have addressed the molecular evolution of HPAIV in Bangladesh and reported the introduction of three distinct clades (clade 2.2, clade 2.3.4 and clade 2.3.2.1) of H5N1 HPAIV (Islam et al., Citation2012; Haque et al., Citation2014). However, pathological investigations of these outbreaks are limited (Khan et al., Citation2014; Haider et al., Citation2017). Here, we report the pathology of HPAI in ducks and quails, as well as immunohistochemical localization of the virus antigen and phylogenetic characterization of the virus from three natural outbreaks.
Materials and methods
Outbreaks
Two carcases of 6-month-old Japanese quails from a commercial farm of Gouripur (of Mymensingh district) and three 2-month-old sick ducks of Khaki Campbell breed from two free-range flocks of Atpara (of Netrokona district) and Fulpur (of Mymensingh district) were submitted to the Department of Pathology, Bangladesh Agricultural University, Mymensingh, in April and July of 2011, respectively. A brief description of the three outbreaks is presented in . The quail farm lost all of its 860 birds within a period of two days; birds died suddenly and often with convulsions. The farmer brought the last two carcases to the Department of Pathology for diagnosis. On the other hand, the ducks were submitted alive with marked nervous signs including in-coordination, torticollis and head tremors. As reported by the farmers, affected ducks suffered for several days before they died and the mortality in ducks reached up to 10% at the time of investigation. The two submitted sick ducks were sacrificed in the laboratory for further investigation.
Table 1. Information about outbreaks investigated in the present study.
Necropsy, sample collection and histopathology
The carcases were subjected to routine necropsy with appropriate biosafety precautions. External and internal gross changes were recorded. Tissue samples from trachea and lungs were also collected aseptically during necropsy for virological investigation. Tissue samples from different organs were collected in 10% neutral buffered formalin for histopathological study. Tissue was processed for paraffin embedding, sectioned and stained with haematoxylin and eosin following standard procedures (Luna, Citation1968).
Immunohistochemistry
Immunohistochemical staining was done on formalin-fixed tissues to detect avian influenza virus antigen. Type A influenza virus nucleoprotein-specific monoclonal antibodies (AbD Serotec, Oxford, UK) and ChemMate TM Detection kit (DAKO, Glostrup, Denmark) were used for the detection of antigen as per the manufacturer’s instructions. Briefly, 4 μm thick sections were mounted on poly-L-lysine coated slides (Thermoshandon, Waltham, MA, USA). After deparaffinization, the fixed antigen was unmasked with 0.1% trypsin (Sigma-Aldrich) and endogenous tissue peroxidase was inactivated with 0.3% hydrogen peroxide. Influenza A virus-specific mouse monoclonal antibody at a dilution of 1:250 was added to the slides and incubated overnight at 4°C in a moist chamber. Then biotinylated goat anti-mouse IgG and streptavidin-horseradish peroxidase conjugate were added sequentially to the slides. Finally, the reaction was developed by adding ready-to-use AEC chromogen and H2O2 substrate. The sections were counterstained with Mayer’s haematoxylin and mounted with glycerol gelatin (Aquaperm, Thermo Shandon, USA). Parallel negative and known positive controls were run each time. Positive tissue sections, incubated with PBS instead of the primary antibody, served as the negative control.
Molecular detection and characterization of virus
RNA was extracted from bird-wise pooled respiratory tissuess, and RT–PCR for M, H5, H9 and N1 genes of AIV and F gene of Newcastle disease virus were performed as described earlier (Fouchier et al., Citation2000; Lee et al., Citation2001; WHO, Citation2007; Barman et al., Citation2017). Partial H5 gene fragments of 545 bp amplified from two separate runs of each sample were sequenced directly from both directions using the PCR primers. The raw sequence data were first checked for quality and then edited and assembled with the Bioedit and MEGA7 (www.megasoftware.net) software. The established HA gene sequences were submitted to GenBank. The GenBank accession numbers are JQ609543, JQ609544 and JN679068 for D-1, D-3 and Q-2 isolates, respectively. In addition, the full length sequences of five internal genes (HA, NA, M, NS and NP) of one duck isolate (D-1) were also generated using primers and protocols of Hoffmann et al. (Citation2001). The HA gene sequences were subjected to a phylogenetic analysis along with related sequences representing each clade downloaded from the GenBank and GISAID databases. Multiple alignment was performed with Clustal W algorithm and a Maximum Likelihood phylogenetic tree was constructed with MEGA7 programme. The stability of the nodes in the phylogenetic trees was tested by bootstrapping with 1000 replications.
Results
Necropsy findings
In quails, generalized congestion in the muscle, haemorrhages, congestion and excess mucus in the larynx and trachea, petechial haemorrhages in the heart, severe congestion in the liver and kidneys and congestion and necrosis in the pancreas were observed at necropsy. Gross lesions in the brain could not be ascertained as the organ was largely autolyzed. On the other hand, no significant gross lesion was found in affected ducks during necropsy, not even in the brain.
Histopathological findings
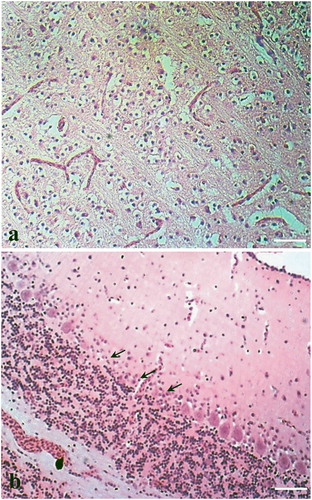
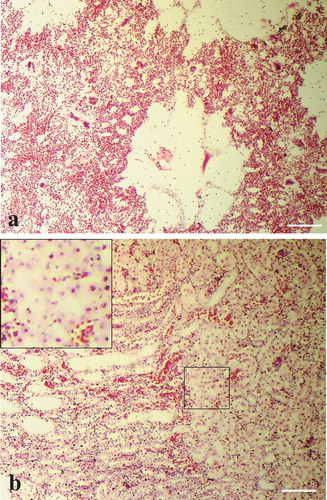
In quails, severe oedema, collapsed alveoli and mononuclear cell infiltration were found in the lung ((a)). Haemorrhages in the cartilage and lamina propria with mononuclear infiltration in adventitia were found in the trachea. Vacuolar degeneration along with sinusoidal congestions, haemorrhages and cellular infiltrations were noticed in the liver. Focal necrosis was seen in acini of the pancreas. The kidney showed tubular necrosis, diffuse congestion and haemorrhages with cellular infiltration in the interstitium ((b)). Haemorrhages and lymphoid depletion in white pulp of the spleen and lymphoid depletion in the bursa of Fabricius were also found. Histopathology of quail brain could not be studied due to autolysis. In the ducks, marked non-purulent encephalitis with neuronal degeneration and gliosis in the brain ((a)) were the most significant histopathological changes. Loss of Purkinje cells in cerebellum was also found ((b)). The liver showed congestions and necrosis with infiltration of inflammatory cells. Changes in other tissues were not prominent.
Figure 1. Histopathological changes in lungs and kidney from avian influenza infected quails (H&E stain, bar = 50 µm). (a) Section of lung showing haemorrhage, collapsed alveoli and mononuclear cell infiltration. (b) Section of kidney showing tubular necrosis, diffuse congestion and haemorrhages with cellular infiltration in the interstitium. Zoomed in view of tubular necrosis in the marked area has been shown in the inset.
Immunohistochemical localization of avian influenza virus antigen
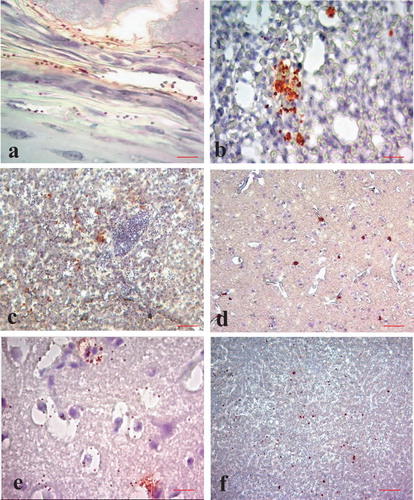
Avian influenza virus antigen localization in quails and ducks was very similar. Virus antigen could be detected in the cells of the mucosa, submucosa and peripheral chondrocytes of the trachea ((a)), interstitial cells in the lung ((b)), hepatocytes in the liver ((c)), degenerating neurons and glial cells in the brain ((d–e)) and degenerating and necrotic cells in pancreatic acini ((f)). Antigen localization in the brain of quails could not be studied due to autolysis.
Figure 3. Immunohistochemical localization of avian influenza virus antigen in different tissues of ducks and quails (IHC counter stained with haematoxylin). Tissue sections from avian influenza infected ducks showing virus antigen in the (a) peripheral chondrocytes (bar = 50 µm), (b) interstitial cells of lungs (bar = 25 µm), (c) cytoplasm of hepatocytes (bar = 50 µm), (d) neurons and glial cells (bar = 50 µm) and (e) degenerating neurons (bar = 50 µm) of brain. (f) Section of pancreas from quail showing virus antigen in the acinar cells of the pancreas (bar = 50 µm).
Molecular detection and phylogenetic analysis
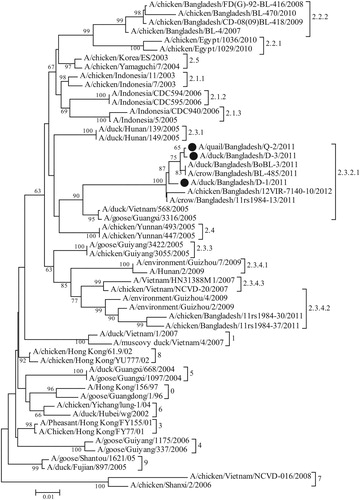
For confirmation of the suspected outbreaks and subtyping of the virus, RT–PCR was performed from the RNA extracted from bird-wise pooled respiratory tissues. In RT–PCR, the H5 and N1 subtype-specific primer sets successfully amplified 545 bp and 343 bp fragments respectively from all three samples, confirming H5N1 subtype of the viruses. RT–PCR of H9 of AIV and F gene of NDV did not produce any positive amplification in these samples, ruling out the chance of co-infections with these viruses. The 545 bp RT–PCR products from the AI suspected quail and duck samples were sequenced. The sequence data of one virus from quail (A/quail/Bangladesh/Q-2/2011) and two viruses from ducks (A/duck/Bangladesh/D-1/2011 and A/duck/Bangladesh/D-3/2011) have been submitted to GenBank (accession numbers: JQ609543, JQ609544 and JN679068). Phylogenetic analysis showed that the three studied Bangladeshi isolates from quail and ducks clustered under clade 2.3.2.1 of H5N1 HPAI virus, together with other HPAI viruses isolated from crows, ducks and chickens during the period of 2011-2012 in Bangladesh (). However, all the previous H5N1 isolates from Bangladesh prior to 2011, and some isolates from 2011, belonged to clade 2.2.2 and two previous isolates belonged to clade 2.3.4.2, indicating at least three separate introductions of H5N1 HPAI viruses into Bangladesh. The full-length sequences of HA, NA, M, NS and NP genes of one of the duck isolates (D1) were also established. The phylogenetic tree based on full-length HA gene sequence (data not shown) was similar to that obtained with partial gene sequences (). The full-length NA, M, NS and NP genes of D1 isolate clustered with that of other clade 2.3.2.1 viruses of 2011 and 2012 in phylogenetic analysis (data not shown).
Figure 4. Maximum Likelihood phylogenetic tree showing the relationship of H5N1 HPAI isolates from quail and ducks from Bangladesh and with the strains representing different clades. The analysis involved 55 nucleotide sequences. There was a total of 533 positions in the final dataset. The evolutionary distances were computed using the Maximum Composite Likelihood method and are in the units of the number of base substitutions per site. Bangladeshi isolates analysed in the present study are indicated by solid circles. Evolutionary analyses were conducted in MEGA7.
Discussion
While all avian species are susceptible to avian influenza viruses, the pathogenicity of the viruses may vary between the species. Wild waterfowls are considered as the reservoir of all avian influenza viruses and they harbour the virus usually without clinical disease (Krauss et al., Citation2007). However, in recent years, H5N1 avian influenza viruses have also caused clinical diseases in ducks but severity varies with the virus strains (Sturm-Ramirez et al., Citation2005; Kim et al., Citation2009). Since the emergence of HPAI in Bangladesh in 2007, the virus has been associated with lethal infections in chickens. However, clinical disease in ducks was not noticed in Bangladesh until 2011. Clinical outbreaks in ducks (Haider et al., Citation2017) coincided with the introduction of a new virus of clade 2.3.2.1 (Islam et al., Citation2012); previously circulating viruses were of clade 2.2. The present isolates also belong to clade 2.3.2.1. The circumstantial evidence suggests that viruses of clade 2.3.2.1 are relatively more virulent for ducks as compared to clade 2.2. Clinical manifestations in ducks were mostly the nervous signs, which were remarkably different from that observed in gallinaceous birds. Similar nervous signs in ducks with variable mortality were also observed by others in natural and experimental infections of clade 2.3.2.1 viruses in Bangladesh, India, Indonesia and other Asian countries (Sturm-Ramirez et al., Citation2005; Nagarajan et al., Citation2012; Dharmayanti et al., Citation2014; Haider et al., Citation2017). Haider et al. (Citation2017) reported a 47% mortality rate in ducks from Bangladesh, whereas it was 61% in India (Nagarajan et al., Citation2012). However, several studies detected viruses of clade 2.3.2.1 in apparently healthy ducks from Bangladesh (Biswas et al., Citation2018; Khan et al., Citation2018). In the present study, we found 10% mortality in ducks. It was, however, not clear if the variations in mortality were due to differences in virulence of the virus or breed and age of the ducks.
In the present investigation, Japanese quails appeared to be highly susceptible to HPAI with 100% mortality in a flock within two days. Several studies also reported high susceptibility of quails to HPAI on experimental infection resulting in rapid mortality with or without clinical signs (Jeong et al., Citation2009; Saito et al., Citation2009; Sun et al., Citation2011; Bertran et al., Citation2013, Citation2014). Severe gross and microscopic vascular, necrotic and inflammatory lesions in the respiratory and digestive organs, as well as in the pancreas and kidneys, were characteristics of acute systemic infection in quails. Acute death of quails might have resulted from respiratory failures. On the contrary, ducks showed prominent nervous signs with non-purulent encephalitis but almost no lesions in the visceral organs. Neurological signs have also been reported in quails (Perkins & Swayne, Citation2001; Bertran et al., Citation2013). However, the extent of involvement of the nervous system in quails in the present investigation could not be evaluated because of autolysis.
Viral antigen was detected by immunohistochemical staining in various organs, such as degenerating neurons and proliferating glial cells in the brain (of ducks), in the mucosa of trachea, pneumocytes and reactive cells in the lungs, hepatocytes in the liver and necrosed acini in the pancreas. Widespread localization of avian influenza virus antigen has been reported previously in quails and ducks (Perkins & Swayne, Citation2001; Ellis et al., Citation2004; Bröjer et al., Citation2009; Haider et al., Citation2017).
The present study illustrates the varying clinical and pathological features of HPAI in ducks and quails following natural infection with the same strain of the virus.
Disclosure statement
No potential conflict of interest was reported by the authors.
ORCID
Mohammed Nooruzzaman http://orcid.org/0000-0002-9358-1494
Additional information
Funding
References
- Barman, L.R., Nooruzzaman, M., Sarker, R.D., Rahman, M.T., Saife, M.R.B., Giasuddin, M., Das, B.C., Das, P.M., Chowdhury, E.H. & Islam, M.R. (2017). Phylogenetic analysis of Newcastle disease viruses from Bangladesh suggests continuing evolution of genotype XIII. Archives of Virology, 162, 3177–3182. doi: 10.1007/s00705-017-3479-x
- Bertran, K., Dolz, R., Busquets, N., Gamino, V., Vergara-Alert, J., Chaves, A.J., Ramis, A., Abad, X.F., Höfle, U. & Majó, N. (2013). Pathobiology and transmission of highly and low pathogenic avian influenza viruses in European quail (Coturnix c. Coturnix). Veterinary Research, 44, 23. doi: 10.1186/1297-9716-44-23
- Bertran, K., Dolz, R. & Majó, N. (2014). Pathobiology of avian influenza virus infection in minor gallinaceous species: a review. Avian Pathology, 43, 9–25. doi: 10.1080/03079457.2013.876529
- Biswas, P.K., Giasuddin, M., Chowdhury, P., Barua, H., Debnath, N.C. & Yamage, M. (2018). Incidence of contamination of live bird markets in Bangladesh with influenza A virus and subtypes H5, H7 and H9. Transboundary and Emerging Diseases, 65, 687–695. doi: 10.1111/tbed.12788
- Bröjer, C., Agren, E.O., Uhlhorn, H., Bernodt, K., Mörner, T., Jansson, D.S., Mattsson, R., Zohari, S., Thorén, P., Berg, M. & Gavier-Widén, D. (2009). Pathology of natural highly pathogenic avian influenza H5N1 infection in wild tufted ducks (Aythya fuligula). Journal of Veterinary Diagnostic Investigation, 21, 579–587. doi: 10.1177/104063870902100501
- Cattoli, G., Fusaro, A., Monne, I., Coven, F., Joannis, T., El-Hamid, H.S., Hussein, A.A., Cornelius, C., Amarin, N.M., Mancin, M., Holmes, E.C. & Capua, I. (2011). Evidence for differing evolutionary dynamics of A/H5N1 viruses among countries applying or not applying avian influenza vaccination in poultry. Vaccine, 29, 9368–9375. doi: 10.1016/j.vaccine.2011.09.127
- Dharmayanti, N.L., Hartawan, R., Pudjiatmoko Wibawa, H., Balish, A., Donis, R., Davis, C.T. & Samaan, G. (2014). Genetic characterization of clade 2.3.2.1 avian influenza A(H5N1) viruses, Indonesia, 2012. Emerging Infectious Diseases, 20, 671–674. doi: 10.3201/eid2004.130517
- Ellis, T.M., Bousfield, R.B., Bissett, L.A., Dyeting, K., Luck, G., Tsim, S., Sturm-Ramirez, K., Webster, R., Guan, Y. & Peiris, J. (2004). Investigation of outbreaks of highly pathogenic H5N1 avian influenza in waterfowl and wild birds in Hong Kong in late 2002. Avian Pathology, 33, 492–505. doi: 10.1080/03079450400003601
- Fouchier, R.A., Bestebroer, T.M., Herfst, S., Van Der Kemp, L., Rimmelzwaan, G.F. & Osterhaus, A.D. (2000). Detection of influenza A viruses from different species by PCR amplification of conserved sequences in the matrix gene. Journal of Clinical Microbiology, 38, 4096–4101.
- Fouchier, R.A., Munster, V., Wallensten, A., Bestebroer, T.M., Herfst, S., Smith, D., Rimmelzwaan, G.F., Olsen, B. & Osterhaus, A.D. (2005). Characterization of a novel influenza A virus hemagglutinin subtype (H16) obtained from black-headed gulls. Journal of Virology, 79, 2814–2822. doi: 10.1128/JVI.79.5.2814-2822.2005
- Guan, Y., Poon, L.L., Cheung, C.Y., Ellis, T.M., Lim, W., Lipatov, A.S., Chan, K.H., Sturm-Ramirez, K.M., Cheung, C.L., Leung, Y.H., Yuen, K.Y., Webster, R.G. & Peiris, J.S. (2004). H5N1 influenza: a protean pandemic threat. Proceedings of the National Academy of Sciences of the United States of America, 101, 8156–8161. doi: 10.1073/pnas.0402443101
- Haider, N., Sturm-Ramirez, K., Khan, S.U., Rahman, M.Z., Sarkar, S., Poh, M.K., Shivaprasad, H.L., Kalam, M.A., Paul, S.K., Karmakar, P.C., Balish, A., Chakraborty, A., Mamun, A.A., Mikolon, A.B., Davis, C.T., Rahman, M., Donis, R.O., Heffelfinger, J.D., Luby, S.P. & Zeidner, N. (2017). Unusually high mortality in waterfowl caused by highly pathogenic avian influenza A (H5N1) in Bangladesh. Transboundary and Emerging Disease, 64, 144–156. doi: 10.1111/tbed.12354
- Haque, M.E., Giasuddin, M., Chowdhury, E.H. & Islam, M.R. (2014). Molecular evolution of H5N1 highly pathogenic avian influenza viruses in Bangladesh between 2007 and 2012. Avian Pathology, 43, 183–194. doi: 10.1080/03079457.2014.898244
- Hoffmann, E., Stech, J., Guan, Y., Webster, R.G. & Perez, D.R. (2001). Universal primer set for the full-length amplification of all influenza A viruses. Archives of Virology, 146, 2275–2289. doi: 10.1007/s007050170002
- Islam, M.R., Haque, M.E., Giasuddin, M., Chowdhury, E.H., Samad, M.A., Parvin, R., Nooruzzaman, M., Rahman, M.M. & Monoura, P. (2012). New introduction of clade 2.3.2.1 avian influenza virus (H5N1) into Bangladesh. Transboundary and Emerging Disease, 59, 460–463. doi: 10.1111/j.1865-1682.2011.01297.x
- Jeong, O.M., Kim, M.C., Kim, M.J., Kang, H.M., Kim, H.R., Kim, Y.J., Joh, S.J., Kwon, J.H. & Lee, Y.J. (2009). Experimental infection of chickens, ducks and quails with the highly pathogenic H5N1 avian influenza virus. Journal of Veterinary Science, 10, 53–60. doi: 10.4142/jvs.2009.10.1.53
- Khan, S.U., Berman, L., Haider, N., Gerloff, N., Rahman, M.Z., Shu, B., Rahman, M., Dey, T.K., Davis, T.C., Das, B.C., Balish, A., Islam, A., Teifke, J.P., Zeidner, N., Lindstrom, S., Klimov, A., Donis, R.O., Luby, S.P., Shivaprasad, H.L. & Mikolon, A.B. (2014). Investigating a crow die-off in January-February 2011 during the introduction of a new clade of highly pathogenic avian influenza virus H5N1 into Bangladesh. Archives of Virology, 159, 509–518. doi: 10.1007/s00705-013-1842-0
- Khan, S.U., Gurley, E.S., Gerloff, N., Rahman, M.Z., Simpson, N., Rahman, M., Haider, N., Chowdhury, S., Balish, A., Zaman, R.U., Nasreen, S., Chandra Das, B., Azziz-Baumgartner, E., Sturm-Ramirez, K., Davis, C.T., Donis, R.O. & Luby, S.P. (2018). Avian influenza surveillance in domestic waterfowl and environment of live bird markets in Bangladesh, 2007–2012. Scientific Report, 8, 9396. doi: 10.1038/s41598-018-27515-w
- Kim, J.K., Negovetich, N.J., Forrest, H.L. & Webster, R.G. (2009). Ducks: the “Trojan horses” of H5N1 influenza. Influenza and Other Respiratory Viruses, 3, 121–128. doi: 10.1111/j.1750-2659.2009.00084.x
- Krauss, S., Obert, C.A., Franks, J., Walker, D., Jones, K., Seiler, P., Niles, L., Pryor, S.P., Obenauer, J.C., Naeve, C.W., Widjaja, L., Webby, R.J. & Webster, R.G. (2007). Influenza in migratory birds and evidence of limited intercontinental virus exchange. PLoS Pathogens, 3, e167. doi: 10.1371/journal.ppat.0030167
- Lee, M.S., Chang, P.C., Shien, J.H., Cheng, M.C. & Shieh, H.K. (2001). Identification and subtyping of avian influenza viruses by reverse transcription-PCR. Journal of Virological Methods, 97, 13–22. doi: 10.1016/S0166-0934(01)00301-9
- Luna, L.G. (1968). Manual of histologic staining methods of the Armed Forces Institute of Pathology, 3rd edn. New York: McGraw Hill Book Co.
- Nagarajan, S., Tosh, C., Smith, D.K., Peiris, J.S., Murugkar, H.V., Sridevi, R., Kumar, M., Katare, M., Jain, R., Syed, Z., Behera, P., Cheung, C.L., Khandia, R., Tripathi, S., Guan, Y. & Dubey, S.C. (2012). Avian influenza H5N1 virus of clade 2.3.2 in domestic poultry in India. PLoS One, 7, e31844. doi: 10.1371/journal.pone.0031844
- OIE. (2017, December 15). Update on highly pathogenic avian influenza in animals. World Organization for Animal Health. http://www.oie.int/animal-health-in-the-world/update-on-avian-influenza/avian_influenza/guidelines/RecAIlabtestsAug07.pdf
- Parvin, R., Kamal, A.H., Haque, M.E., Chowdhury, E.H., Giasuddin, M., Islam, M.R. & Vahlenkamp, T.W. (2014). Genetic characterization of highly pathogenic H5N1 avian influenza virus from live migratory birds in Bangladesh. Virus Genes, 49, 438–448. doi: 10.1007/s11262-014-1118-0
- Perkins, L.E. & Swayne, D.E. (2001). Pathobiology of A/Chicken/Hong Kong/220/97 (H5N1) avian influenza virus in seven gallinaceous species. Veterinary Pathology, 38, 149–164. doi: 10.1354/vp.38-2-149
- Saito, T., Watanabe, C., Takemae, N., Chaisingh, A., Uchida, Y., Buranathai, C., Suzuki, H., Okamatsu, M., Imada, T., Parchariyanon, S., Traiwanatam, N. & Yamaguchi, S. (2009). Pathogenicity of highly pathogenic avian influenza viruses of H5N1 subtype isolated in Thailand for different poultry species. Veterinary Microbiology, 133, 65–74. doi: 10.1016/j.vetmic.2008.06.020
- Sarker, R.D., Giasuddin, M., Chowdhury, E.H. & Islam, M.R. (2017). Serological and virological surveillance of avian influenza virus in domestic ducks of the north-east region of Bangladesh. BMC Veterinary Research, 13, 180. doi: 10.1186/s12917-017-1104-6
- Sturm-Ramirez, K.M., Hulse-Post, D.J., Govorkova, E.A., Humberd, J., Seiler, P., Puthavathana, P., Buranathai, C., Nguyen, T.D., Chaisingh, A., Long, H.T., Naipospos, T.S., Chen, H., Ellis, T.M., Guan, Y., Peiris, J.S. & Webster, R.G. (2005). Are ducks contributing to the endemicity of highly pathogenic H5N1 influenza virus in Asia? Journal of Virology, 79, 11269–11279. doi: 10.1128/JVI.79.17.11269-11279.2005
- Sun, H., Jiao, P., Jia, B., Xu, C., Wei, L., Shan, F., Luo, K., Xin, C., Zhang, K. & Liao, M. (2011). Pathogenicity in quails and mice of H5N1 highly pathogenic avian influenza viruses isolated from ducks. Veterinary Microbiology, 152, 258–265. doi: 10.1016/j.vetmic.2011.05.009
- Webster, R.G., Bean, W.J., Gorman, O.T., Chambers, T.M. & Kawaoka, Y. (1992). Evolution and ecology of influenza A viruses. Microbiological Reviews, 56, 152–179.
- WHO. (2007, August). Recommendations and laboratory procedures for detection of avian influenza A (H5N1) virus in specimens from suspected human cases. World Health Organization. http://www.who.int/csr/disease/
- World Health Organization (WHO). (2014). Revised and updated nomenclature for highly pathogenic avian influenza A (H5N1) viruses. Influenza and Other Respiratory Viruses, 8, 384–388. doi: 10.1111/irv.12230